m5C Modifications in RNA and Cancer
Explore how the epigenetic modification m5C regulates the fate of RNA molecules and cancer prognosis.
5-methylcytosine (m5C) is an epigenetic modification that involves the addition of a methyl group (CH3) to the carbon-5 position of the cytosine base in nucleic acids. Dynamic, reversible, and widely distributed, m5C modifications can affect the fate of many RNA molecules.
Effect of m5C modifications on RNA
tRNA
In tRNA, m5C modifications promote base pairing, control translation efficiency and accuracy, regulate tRNA stability, and participate in cellular metabolism and stress responses [2]. Methylation of the C38 locus on the anticodon loop in mouse tRNA-Asp can promote the translation of multiple Asp proteins and protect tRNA from stress-induced endonuclease-mediated cleavage [3]. Methylation of the C48 site enhances the hydrophobicity of base pairs and promotes base stacking to enhance protein synthesis [4].
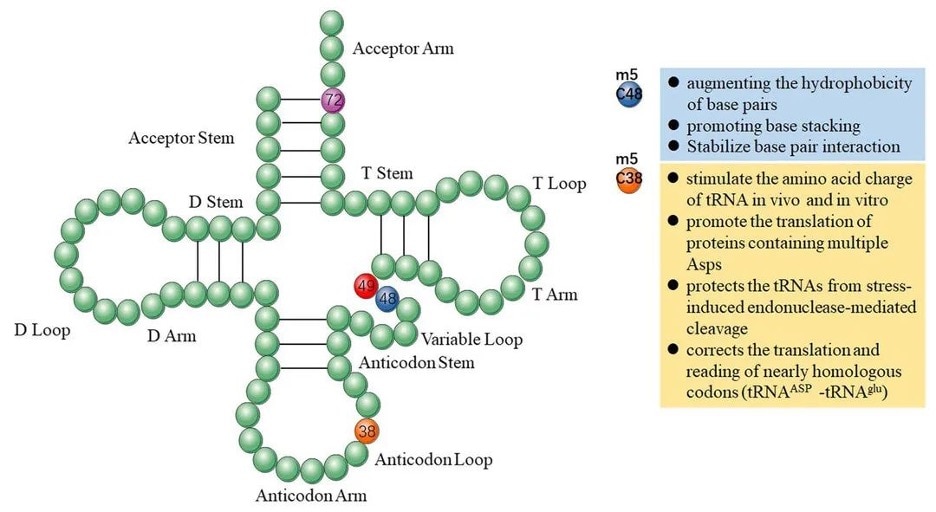
Figure 1 Functions of m5C in tRNA originating. Image from Song et al., 2022 [2]
rRNA
In rRNA, m5C modifications help keep its conformation stable. Under oxidative stress conditions, rRNA m5C modifications in yeast facilitate rRNA folding and promote selective recruitment and translation of mRNA to participate in different cell signaling pathways [5].
lncRNA
Researchers have also studied the distribution and regulation of m5C on lncRNA and found that m5C is positively correlated with lnc RNA transcription. It is also involved in long-distance chromatin interactions at lncRNA sites. Additionally, lncRNAs regulated by different m5C regulators are associated with the presence of tumors [6].
mRNA
Previously, due to the low abundance of mRNA and the lack of effective isolation and purification techniques, relatively few studies were conducted. However, with the advancement of detection methods in recent years, the research on mRNA m5C modifications has gradually increased.
In mRNA, m5C modifications involve a variety of enzymes and effector molecules, including NOP2/SUN RNA methyltransferase 2 (NSUN2), NSUN6, tRNA aspartate methyltransferase 1 (TRDMT1), and Aly/REF output factor (ALYREF) [7]. The functions of m5C modifications in mRNA include promoting nucleocytoplasmic transport, viral protein expression, DNA repair, mRNA stabilization, proliferation and migration, development, and mRNA splicing regulation.
The distribution of m5C in different cell types or specific locations of mRNA shows unique regulatory activities, which can promote or inhibit translation [1]. Therefore, abnormal mRNA m5C modifications are strongly associated with the development of a variety of diseases, including cancer and autoimmune diseases [8].
m5C regulators
Methylation of RNA mainly involves three types of regulators: 1) Writers that are responsible for adding specific chemical groups to RNA; 2) Readers of RNA modifications to maintain RNA stability and participate in RNA translation and splicing; 3) Erasers that are responsible for removing the modification.
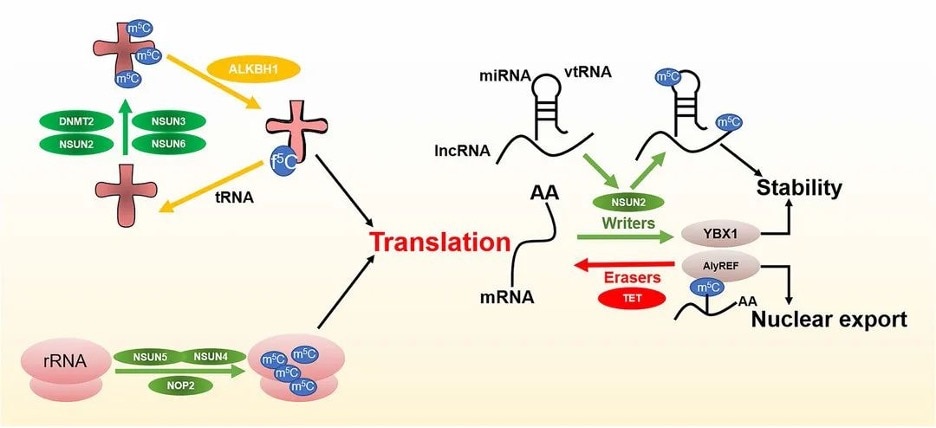
Figure 2 m5C regulators and functions. Image from Song et al., 2022 [2]
m5C writers
m5C writers or methyltransferases use adenosylmethionine as a methyl donor to form m5C by transferring the methyl group to a cytosine.
Several RNA methyltransferases for m5C have been discovered, including DNA methyltransferase 1 and 2 (DNMT1, DNMT2), the tRNA-specific methyltransferase (TRDMT) family, and the NOL1/NOP2/SUN domain (NSUN) protein family.
NSUN2 has a relatively wide range of targets and plays a major role in the methylation of mRNAs, miRNAs, and tRNAs [9]. TRDMT1 catalyzes m5C modifications in tRNA, but unlike NSUN2, methylation by TRDMT1 occurs only at a single cysteine site.
m5C readers
Biological processes in response to m5C modifications require specific proteins (known as readers) to recognize and bind to modification sites. At present, the research on m5C readers is still in the early stages.
ALYREF is an mRNA-reading protein found in the nucleus and plays a role in the nucleoplasmic output of mRNA [10]. YBX1 has also been identified as an mRNA m5C recognition protein in the cytoplasm and maintains the stability of m5C-modified mRNA [11]. In addition, RAD52 has been observed to have a high affinity for hybrid strands of m5C-modified RNA and DNA, indicating that it is an m5C reader of DNA damage sites.
m5C erasers
Modifications, including methylation, are reversible and often are dynamically regulated. However, the classification and definition of demethylases (erasers) for m5C are not well understood.
Early studies identified a demethylation pathway associated with m5C modifications in DNA. TETs, acting as dioxygenases, can oxidize m5C to 5caC (5-carboxycystine). 5caC is then cleaved by thymine-DNA glycosylase, completing the DNA demethylation modification.
Fu et al. found a similar process on RNA where TETs catalyzed the conversion of m5C to hm5C by α-ketoglutaric acid and iron ions. hm5C was further converted to 5fC (5-fluorocytosine) and 5caC by TETs, and finally, the site was converted to cytosine by a base excision repair pathway resulting in demethylation [12].
In addition, another DNA/RNA dioxygenase, ALKBH1, has also been found to be involved in the demethylation of tRNA [13]. Deletion of ALKBH1 has led to severe deficiencies in mitochondrial translation and oxygen consumption, suggesting that m5C may regulate mitochondrial activity [14].
m5C modifications and cancer
Studies in recent years have found that m5C modifications in RNA are associated with a variety of diseases, especially cancers.
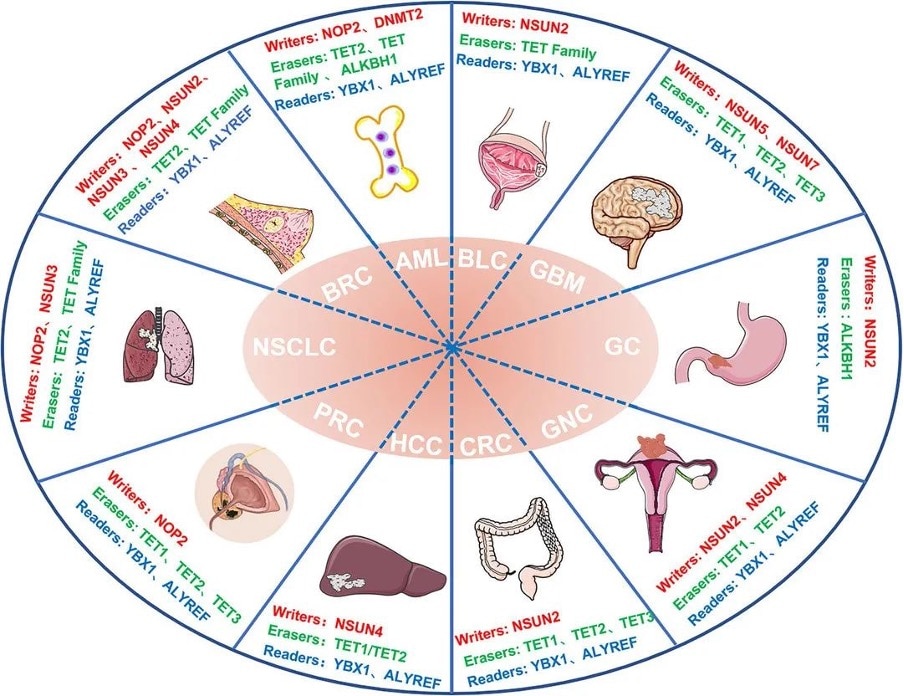
Figure 3 Abnormalities in m5C regulators in cancer. Image from Song et al., 2022 [2] NSCLS: Non-Small Cell Lung Cancer; BRC: Breast Cancer; AML: Acute myeloid leukemia; BLC: Bladder Cancer; GBM: Glioblastoma; GC: Gastric Cancer; GNC: Gynecologic Cancer; CRC: Colorectal Cancer; HCC: Hepatocellular carcinoma; PRC: Prostate Cancer; HDFs: Human diploid fibroblasts; U87: Human glioma cell line U87; ATX: Autotaxin
m5C methylation in mRNA in liver cancer tissues are significantly higher than controls and show unique gene distribution [15]. A study also showed that high expression of NSUN4 and ALYREF is associated with poor prognosis in patients with hepatocellular carcinoma, although the specific underlying mechanism is unclear [16].
Higher expression of the writer NSUN2 is observed in gastric cancer and this may promote the proliferation of gastric cancer cells in an m5C-dependent manner by inhibiting the expression of p57, Kip2 [17]. Additionally, the reader YBX1 can recognize m5c-modified mRNA and recruit ELAVL1 to stabilize HDGF mRNA, ultimately promoting the proliferation and metastasis of bladder cancer cells [18].
Other studies have shown that patients with papillary thyroid carcinoma (PTC) with low m5C modification levels have higher CD4+ memory T cells and CD8+ T cells in the resting state and also have a better prognosis. In patients with PTC with high m5C modifications, activated NK cells and monocytes are mostly abundant and have a poor prognosis [19].
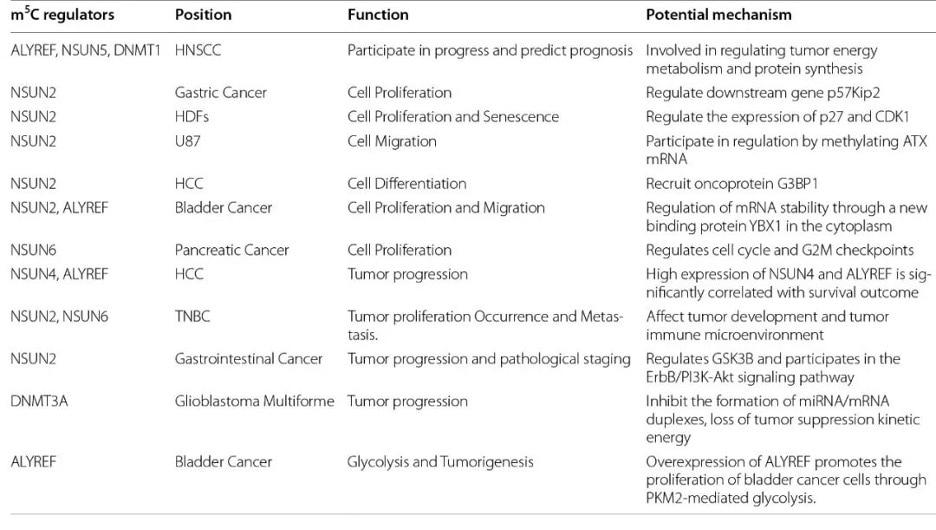
Fig. 4 m5C regulators and cancer. Figure from Song et al., 2022 [2]
As m5C modifications continue to be studied, especially in the context of cancer, scientists are discovering new targets and directions for cancer diagnosis, treatment, and prognosis.
Explore our complete antibody portfolio for m5C modifications
|
m5C Monoclonal Antibody (68301-1-Ig)
|
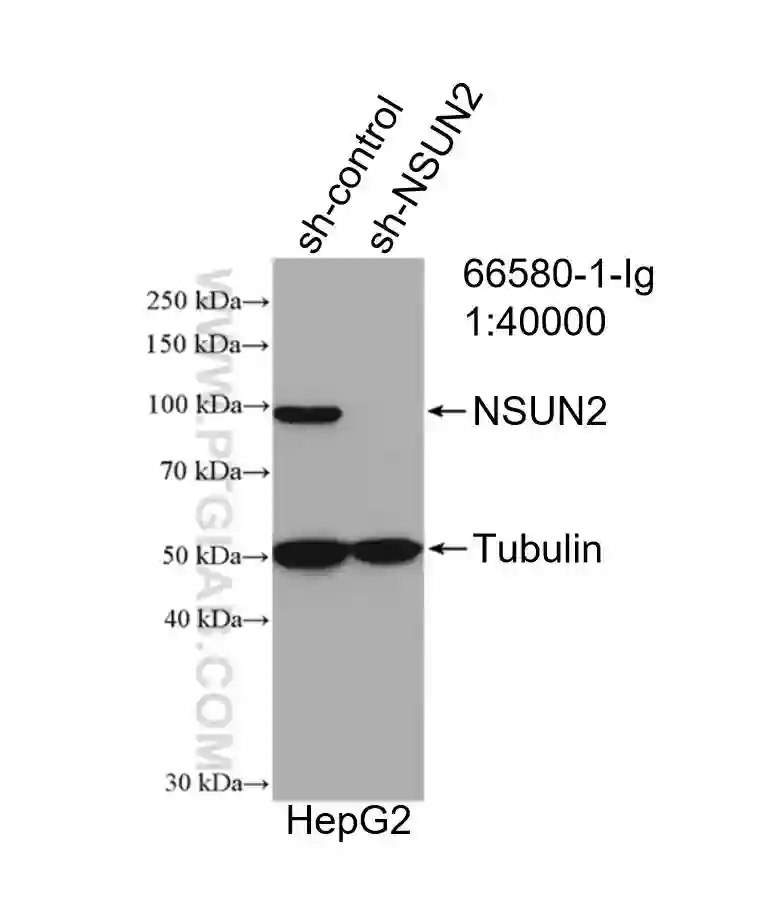
NSUN2 Monoclonal Antibody (66580-1-Ig)
WB result of NSUN2 antibody (66580-1-Ig; 1:40000) with negative control and NSUN2 overexpressed HepG2 cells..
|
|
Target |
Catalog No. |
Host/Type |
Applications |
|
68301-1-Ig |
Mouse monoclonal |
IHC, Dot blot, ELISA |
|
|
16690-1-AP |
Rabbit polyclonal |
WB, IHC, IF, ELISA |
|
|
24206-1-AP |
Rabbit polyclonal |
WB, IP, IHC, IF, CoIP, ChIP, ELISA |
|
|
68485-1-Ig |
Mouse monoclonal |
WB, ELISA |
|
|
19221-1-AP |
Rabbit polyclonal |
WB, ELISA |
|
|
20954-1-AP |
Rabbit polyclonal |
WB, RIP, IP, IHC, IF, ChIP, ELISA |
|
|
26971-1-AP |
Rabbit polyclonal |
WB, IHC, IF, CoIP, ChIP, ELISA |
|
|
14939-1-AP |
Rabbit polyclonal |
WB, ELISA |
|
|
60064-1-Ig |
Mouse monoclonal |
WB, IHC, IF, CoIP, ELISA |
|
|
11910-1-AP |
Rabbit polyclonal |
WB, RIP, IP, IHC, IF, FC, CoIP, CLIP, ChIP, ELISA |
|
|
20854-1-AP |
Rabbit polyclonal |
WB, IP, IHC, IF, FC, ELISA |
|
|
66580-1-Ig |
Mouse monoclonal |
WB, IP, IHC, IF, ELISA |
|
|
15449-1-AP |
Rabbit polyclonal |
WB, IHC, IF, ELISA |
|
|
17240-1-AP |
Rabbit polyclonal |
WB, RIP, IHC, IF, ELISA |
|
|
17546-1-AP |
Rabbit polyclonal |
WB, IHC, ELISA |
|
|
67024-1-Ig |
Mouse monoclonal |
WB, IHC, IF, ELISA |
|
|
28045-1-AP |
Rabbit polyclonal |
WB, IP, ELISA |
|
|
21207-1-AP |
Rabbit polyclonal |
WB, IP, IHC, IF, FC, CoIP, ChIP, ELISA |
|
|
20339-1-AP |
Rabbit polyclonal |
WB, RIP, IHC, IF, CoIP, ChIP, ELISA |
References:
1. Guo G, Pan K, Fang S, Ye L, Tong X, Wang Z, Xue X, Zhang H. Advances in mRNA 5-methylcytosine modifications: Detection, effectors, biological functions, and clinical relevance. Mol Ther Nucleic Acids. 2021 Aug 26; 26:575-593.
2. Song H, Zhang J, Liu B, Xu J, Cai B, Yang H, Straube J, Yu X, Ma T. Biological roles of RNA m5C modification and its implications in Cancer immunotherapy. Biomark Res. 2022 Apr 1; 10(1):15.
3.Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006 Jan 20; 311(5759):395-8.
4.Xu L, Liu X, Sheng N, Oo KS, Liang J, Chionh YH, Xu J, Ye F, Gao YG, Dedon PC, Fu XY. Three distinct 3-methylcytidine (m3C) methyltransferases modify tRNA and mRNA in mice and humans. J Biol Chem. 2017 Sep 1; 292(35):14695-14703.
5.Sharma S, Yang J, Watzinger P, Kötter P, Entian KD. Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res. 2013 Oct; 41(19):9062-76.
6.Hu H, Shu M, He L, Yu X, Liu X, Lu Y, Chen Y, Miao X, Chen X. Epigenomic landscape of 5-hydroxymethylcytosine reveals its transcriptional regulation of lncRNAs in colorectal cancer. Br J Cancer. 2017 Feb 28; 116(5):658-668.
7.Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY, Li A, Wang X, Bhattarai DP, Xiao W, Sun HY, Zhu Q, Ma HL, Adhikari S, Sun M, Hao YJ, Zhang B, Huang CM, Huang N, Jiang GB, Zhao YL, Wang HL, Sun YP, Yang YG. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 2017 May; 27(5):606-625.
8.Luo Y, Feng J, Xu Q, Wang W, Wang X. NSun2 Deficiency Protects Endothelium From Inflammation via mRNA Methylation of ICAM-1. Circ Res. 2016 Mar 18; 118(6):944-56.
9.Khoddami, V. and Cairns, B.R. (2014) Transcriptome-Wide Target Profil-ing of RNA Cytosine Methyltransferases Using the Mechanism-Based Enrichment Procedure Aza-IP. Nature Protocols, 9, 337-361.
10.Li W, Xu L. Epigenetic Function of TET Family, 5-Methylcytosine, and 5-Hydroxymethylcytosine in Hematologic Malignancies. Oncol Res Treat. 2019; 42(6):309-318.
11.Takai, H., Masuda, K., Sato, T., et al. (2014) 5-Hydroxymethylcytosine Plays a Critical Role in Glioblastomagenesis by Recruiting the CHTOP-Methylosome Com-plex. Cell Reports, 9, 48-60.
12.Tuorto F, Herbst F, Alerasool N, Bender S, Popp O, Federico G, Reitter S, Liebers R, Stoecklin G, Gröne HJ, Dittmar G, Glimm H, Lyko F. The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J. 2015 Sep 14; 34(18):2350-62.
13.Haag S, Sloan KE, Ranjan N, et 0al. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J. 2016; 35(19):2104–19
14.Ougland R, Jonson I, Moen MN, Nesse G, Asker G, Klungland A, Larsen E. Role of ALKBH1 in the Core Transcriptional Network of Embryonic Stem Cells. Cell Physiol Biochem. 2016; 38(1):173-84.
15. Zhang Q, Zheng Q, Yu X, He Y, Guo W. Overview of distinct 5-methylcytosine profiles of messenger RNA in human hepatocellular carcinoma and paired adjacent non-tumor tissues. J Transl Med. 2020 Jun 22; 18(1):245.
16. He Y, Yu X, Li J, Zhang Q, Zheng Q, Guo W. Role of m5C-related regulatory genes in the diagnosis and prognosis of hepatocellular carcinoma. Am J Transl Res. 2020 Mar 15; 12(3):912-922.
17.Mei L, Shen C, Miao R, Wang JZ, Cao MD, Zhang YS, Shi LH, Zhao GH, Wang MH, Wu LS, Wei JF. RNA methyltransferase NSUN2 promotes gastric cancer cell proliferation by repressing p57Kip2 in an m5C-dependent manner. Cell Death Dis. 2020 Apr 24; 11(4):270.
18. Chen X, Li A, Sun BF, Yang Y, Han YN, Yuan X, Chen RX, Wei WS, Liu Y, Gao CC, Chen YS, Zhang M, Ma XD, Liu ZW, Luo JH, Lyu C, Wang HL, Ma J, Zhao YL, Zhou FJ, Huang Y, Xie D, Yang YG. 5-methylcytosine promotes the pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol. 2019 Aug; 21(8):978-990.
19. Li F, Deng Q, Pang X, et al. M (5) C regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in papillary thyroid carcinoma. Front Oncol. 2021; 11:729887
Related Content
Detecting RNA Methylation by Dot Blotting
RIP/RIP-seq to study RNA Modifications
Support
Newsletter Signup
Stay up-to-date with our latest news and events. New to Proteintech? Get 10% off your first order when you sign up.
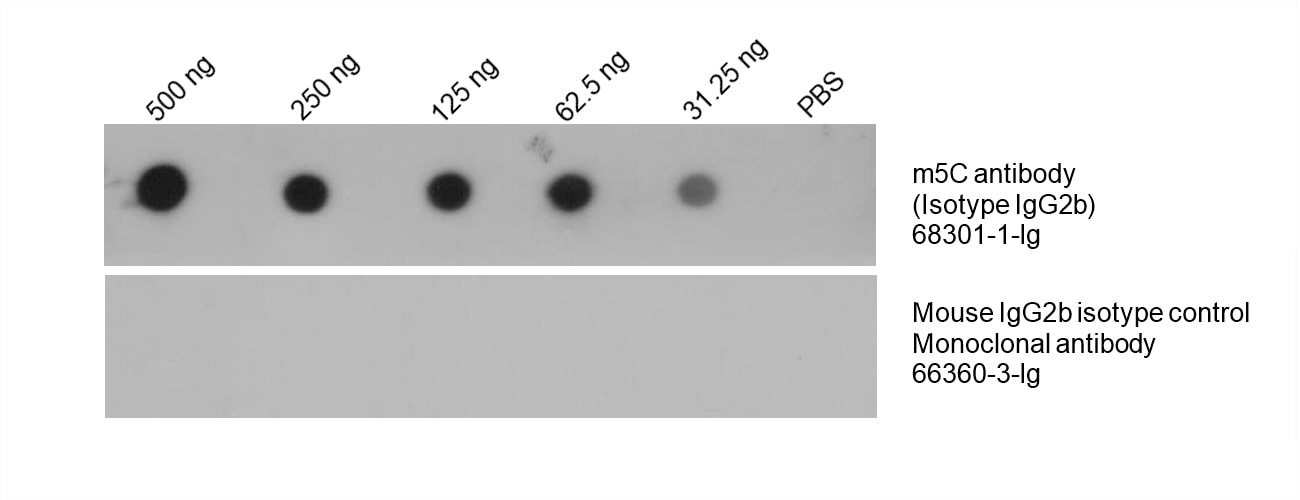 Total DNA was isolated from HeLa cells and dotted to an NC membrane at different amounts. The membrane was blotted with m5C antibody (68301-1-Ig, 1:5000). A parallel dot blot was performed using Mouse IgG2b isotype control Monoclonal antibody 66360-3-Ig at the same dose.
Total DNA was isolated from HeLa cells and dotted to an NC membrane at different amounts. The membrane was blotted with m5C antibody (68301-1-Ig, 1:5000). A parallel dot blot was performed using Mouse IgG2b isotype control Monoclonal antibody 66360-3-Ig at the same dose.