Unlocking the Secrets of Aging: The Role of Epigenetics in Cellular Senescence
Written by Shiksha Dutta, PhD student at University of Montreal
What is cellular senescence?
Aging is an inevitable part of life, but what if we could understand its mechanisms better? Enter cellular senescence, a critical process where cells stop dividing and undergo significant changes in response to various cellular stressors, including genotoxic stress or DNA insults (Fig 1). Cellular senescence acts as a crucial tumor suppression mechanism by limiting the multiplication of cells by undergoing senescence-associated proliferation arrest (SAPA) and activating the immune response. It also maintains tissue homeostasis and wound healing through tissue remodeling mechanisms. However, the accumulation of senescent cells that is associated with aging is primarily associated with cancer progression and age-associated organ pathologies. Therefore, while cellular senescence acts as a cancer suppressing mechanism initially, the accumulation of these senescent cells can contribute to cancer, chronic inflammation, aging, and age-related diseases.
Cellular senescence is characterized by a series of autocrine and paracrine proinflammatory factors, which form key components of the senescence-associated secretory phenotype (SASP). The SASP remodels the tissue microenvironment and contributes to the accumulation of senescent cells associated with aging. These include soluble signaling factors such as interleukins (IL-6, -7, -13, -15), chemokines (IL-8, MCP-2, -4, -1), growth factors (VEGF, IGFBP-2, -3, -4, -6, -7), secretion of proteases (MMP-1, -3, -10, -12, -13, -14), and changes in the extracellular matrix. Additionally, senescence is a consequence of DNA double-strand breaks (DSBs) caused by genotoxic stress, including cytotoxic drugs, oxidation, irradiation and oncogene-induced stress. Key players in this biological process are tumor suppressor pathways like p53/p21 and p16INK4a/Rb, which inhibit kinases that regulate cell cycle progression and proliferation. Epigenetic modifications function as puppeteers, controlling how these pathways are activated or silenced in the context of aging. Therefore, the journey from a dividing cell to a senescent one is guided by epigenetic roadmaps.
As well as being induced by external factors such as cytotoxic drugs and irradiation, senescence is also caused by the shortening of telomeres (the protective caps on the end of chromosomes). As well as being influenced by telomerase dysfunction, telomere shortening is an unavoidable consequence of the repeated cellular division seen in aging. While telomerase vastly improves the conservation of telomere length when replicating in the 5’ to 3’ direction, the process is not perfect, and base pairs are lost with every division. The rate at which telomere shortening occurs is influenced by a wide variety of epigenetic factors including socio-economic status, stress and smoking status. When telomere length reaches a level where DNA protection can no longer occur, the cell will either continue to divide and risk becoming cancerous, undergo apoptosis, or enter a senescent state. As well as other, cellular stress-based pathways, this pathway for cellular senescence induction contributes significantly to the accumulation of senescent cells associated with aging.
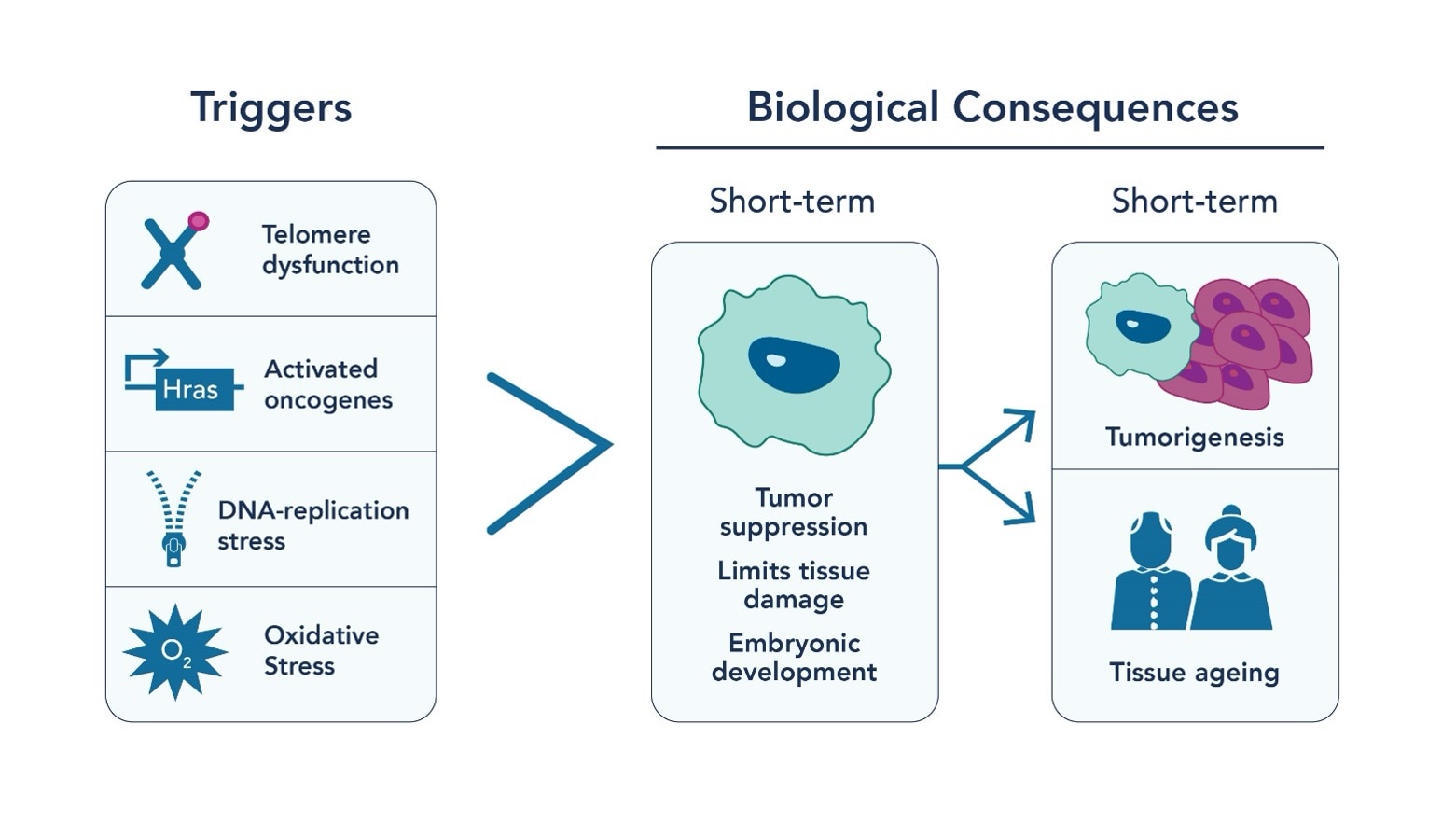
Fig. 1 Triggers for cellular senescence
How epigenetics regulates cellular senescence
Histone Modifications:
Epigenetics investigates changes in gene function that are not driven by modifications in the DNA sequence. It controls when and how genes are expressed or “turned on” through mechanisms like histone modifications and non-coding RNA. Chromatin structure refers to the organization of DNA and proteins within the nucleus of a cell, which allows for the efficient packaging of DNA into a compact form. This structure plays a crucial role in regulating gene expression, DNA replication, and repair. Histones are the main proteins involved in chromatin structure, acting as beads around which DNA winds, thereby organizing it into units called nucleosomes. There are five main types of histones: H1, H2A, H2B, H3, and H4. Two each of H2A, H2B, H3, and H4 form a core around which DNA wraps, while H1 is involved in the higher order packing of these nucleosomes. This arrangement not only compacts the DNA but also plays a key role in the accessibility of specific DNA regions for transcription and other DNA-related processes.
Recent studies have shed light on the epigenetic landscape of senescent cells (Fig 2), offering insights into aging and potential therapeutic targets. Researchers have identified multiple histone post-translational modifications (PTMs) like acetylation and methylation to be essential for senescence associated gene expression. For example, H3K27 acetylation and bromodomain-containing protein 4 (BRD4) recruitment are necessary for the expression of SASP-coding genes. Inhibiting BRD4 can suppress SASP expression and affect immune response. Similarly, H3K4 methyltransferase mixed-lineage leukemia 1 (MLL1) and DOT1L are essential for SASP activation. It is well-studied that a decrease in DNA methylation is observed in senescent cells and is caused by a reduction in DNA methyltransferase 1 (DNMT1). Altered methylation patterns are seen in replicative senescence (senescence caused by telomere shortening associated with cellular division), especially at immune response genes. Furthermore, DNA methylation patterns can predict chronological age by analyzing specific sites on the DNA where methyl groups are added or removed over time. These sites, known as CpG sites, undergo methylation changes that correlate with an organism's age. The methylation profile of senescent cells shares similarities with aging and cancer cells, but there are distinct differences in specific CpG sites.
Chromatin Structure:
The epigenetic landscape of senescent cells also indicates alterations in chromatin structure, particularly the loss and instability of heterochromatin. This change leads to increased transcriptional activity of genes that were previously repressed. In proliferating cells, chromatin modifiers such as polycomb repressor complexes (PRC) suppress genes associated with senescence. Moreover, the interplay between non-coding RNA, such as PANDA, and nuclear transcription factor Y subunit alpha (NF-YA) is instrumental in regulating senescence. Senescence-associated heterochromatin foci (SAHF) represent another key epigenetic feature in senescent cells. These specialized heterochromatin regions silence genes that promote cell proliferation. The formation of SAHF is influenced not only by histone variants but also by the cell type and the method of senescence induction. Histone variants such as H3.3, macroH2A, H2A.J, and H2A.Z are notably prevalent in senescent cells. They play critical roles in transcriptional regulation, DNA repair, and the maintenance of epigenetic states, thereby impacting both SAHF formation and SASP gene expression.
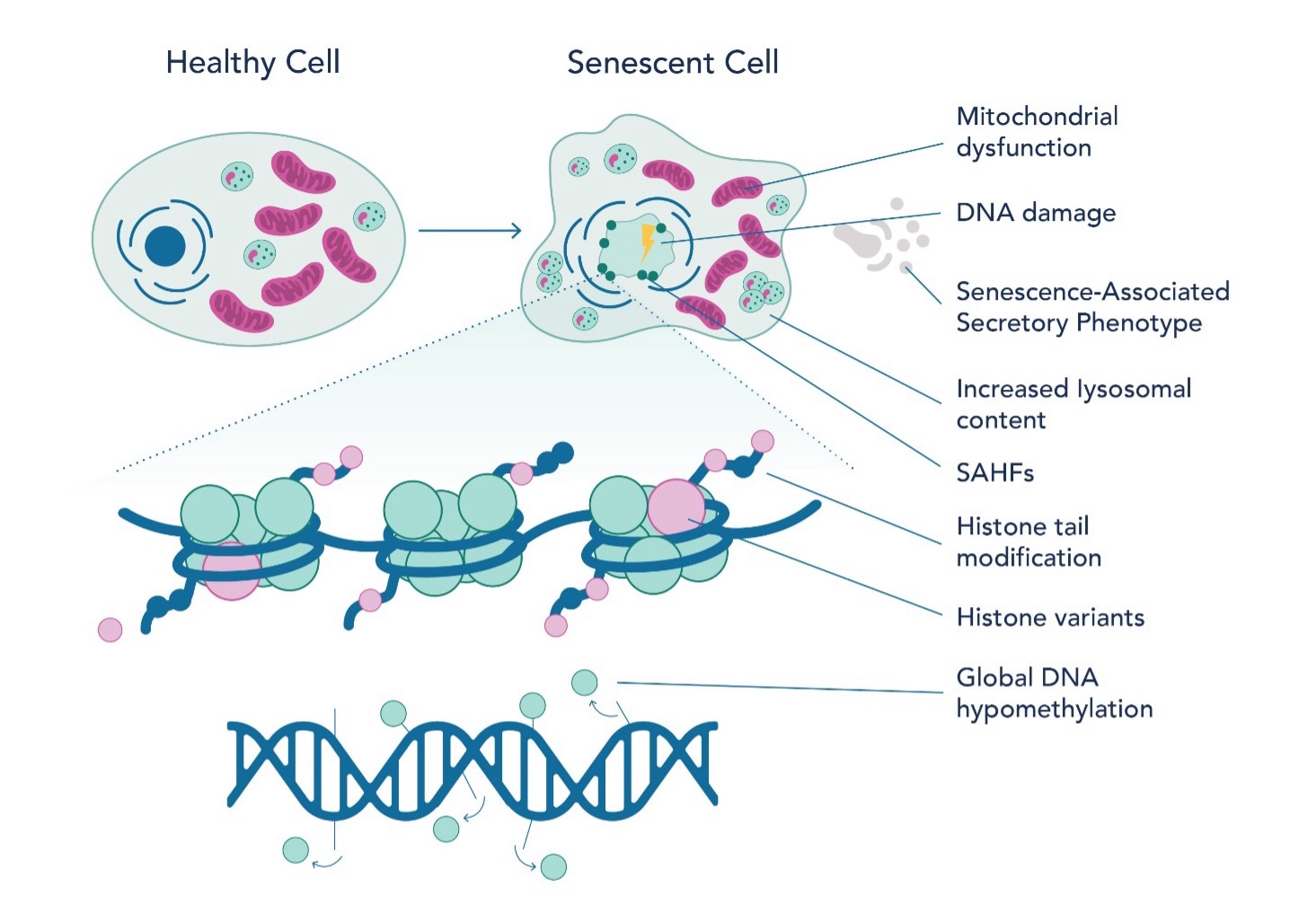
Fig. 2 Epigenetic landscape of senescent cells
In conclusion, exploring epigenetic regulation in cellular senescence has revealed a complex and intricate network of mechanisms that govern the mechanism of aging. From the critical roles of histone modifications and DNA methylation patterns to the dynamic interplay between chromatin structure and non-coding RNA, these findings underscore the profound impact of epigenetic factors on cellular behavior. The insights gained enhance our understanding of cellular aging and open new avenues for therapeutic interventions targeting age-related diseases. As we unravel the mysteries of the epigenome, it becomes increasingly evident that manipulating these epigenetic processes could hold the key to mitigating the effects of aging and improving human health span. The future of aging research lies in harnessing the power of epigenetics, offering promising prospects for developing innovative strategies to combat the challenges of aging.
Antibodies for Epigenetics and Cellular Senescence Targets
|
Target |
Catalog No |
|
Histone H2A.X |
|
|
Histone H2A.Z |
|
|
Histone H2B |
|
|
Histone H3 |
|
|
Acetyl-Histone H3 (lys27) |
|
|
BRD4 |
|
|
MLL |
|
|
DNMT1 |
|
|
NFYA |
|
|
P16-INK4A |
|
|
CDKN2A |
|
|
P21 |
|
|
P53 |
Edited by Ben Raven, PhD student at the University of Sheffield
References:
- Rodier, Francis, and Judith Campisi. "Four faces of cellular senescence." Journal of Cell Biology 192, no. 4 (2011): 547-556.
- Malaquin, Nicolas, Aurélie Martinez, and Francis Rodier. "Keeping the senescence secretome under control: molecular reins on the senescence-associated secretory phenotype." Experimental gerontology 82 (2016): 39-49.
- Pazolli, Ermira, Elise Alspach, Agnieszka Milczarek, Julie Prior, David Piwnica-Worms, and Sheila A. Stewart. "Chromatin Remodeling Underlies the Senescence-Associated Secretory Phenotype of Tumor Stromal Fibroblasts That Supports Cancer Progression Regulation of Stroma-Derived OPN." Cancer research 72, no. 9 (2012): 2251-2261.
- Li, Bing, Michael Carey, and Jerry L. Workman. "The role of chromatin during transcription." Cell 128, no. 4 (2007): 707-719.
- Coppé, Jean-Philippe, Christopher K. Patil, Francis Rodier, Y. U. Sun, Denise P. Muñoz, Joshua Goldstein, Peter S. Nelson, Pierre-Yves Desprez, and Judith Campisi. "Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor." PLoS biology 6, no. 12 (2008): e301.
- De Lera, Angel R., and A. Ganesan. "Two-hit wonders: The expanding universe of multitargeting epigenetic agents." Current Opinion in Chemical Biology 57 (2020): 135-154.
- Sidler, Corinne, Olga Kovalchuk, and Igor Kovalchuk. "Epigenetic regulation of cellular senescence and aging." Frontiers in genetics 8 (2017): 138.
- Astuti Y, Wardhana A, Watkins J, Wulaningsih W; PILAR Research Network. Cigarette smoking and telomere length: A systematic review of 84 studies and meta-analysis. Environ Res. 2017 Oct;158:480-489. doi: 10.1016/j.envres.2017.06.038. Epub 2017 Jul 10. PMID: 28704792; PMCID: PMC5562268.
- Alexeeff SE, Schaefer CA, Kvale MN, Shan J, Blackburn EH, Risch N, Ranatunga DK, Jorgenson E, Hoffmann TJ, Sakoda LC, Quesenberry CP, Van Den Eeden SK. Telomere length and socioeconomic status at neighborhood and individual levels among 80,000 adults in the Genetic Epidemiology Research on Adult Health and Aging cohort. Environ Epidemiol. 2019 May 1;3(3):e049. doi: 10.1097/EE9.0000000000000049. PMID: 33778338; PMCID: PMC7939422.
- Mathur MB, Epel E, Kind S, Desai M, Parks CG, Sandler DP, Khazeni N. Perceived stress and telomere length: A systematic review, meta-analysis, and methodologic considerations for advancing the field. Brain Behav Immun. 2016 May;54:158-169. doi: 10.1016/j.bbi.2016.02.002. Epub 2016 Feb 4. PMID: 26853993; PMCID: PMC5590630.
Related Content
m5C Modifications in RNA and Cancer
RIP/RIP-seq to study RNA Modifications
Detecting RNA Methylation by Dot Blotting
7 Tech Tips For Successful ChIP Experiments
Support
Newsletter Signup
Stay up-to-date with our latest news and events. New to Proteintech? Get 10% off your first order when you sign up.
