ChromoTek’s Nanobody helps LMU scientists in their quest to understand a giant protein complex
A collaboration between ChromoTek GmbH and Ludwig-Maximilians-University Munich (Germany) has yielded a novel nanobody that sheds new light on an important protein complex called INO80.
A collaboration between ChromoTek GmbH and Ludwig-Maximilians-University Munich (Germany) has yielded a novel nanobody VHHs that sheds new light on an important protein complex called INO80. This work was recently published in the high-impact journal Nature Structural & Molecular Biology (Knoll et al. 2018).
Camelid single-domain antibodies (VHHs) have become a popular tool for biochemists and structural biologists in recent years. Like conventional antibodies, Nanobody VHH can inhibit or activate a protein and trap it in a desired conformation (e.g. the activated state). This conformational trapping is handy both for biochemical assays and structure determination. However, nanobodies are tiny single-domain antibodies and are thus able to reach all those interesting spots hidden away in the protein’s clefts and pockets, from which comparatively huge conventional antibodies are excluded. In consequence, nanobodies have emerged as the antibody class of choice if you want to study your favourite protein using biochemical assays, protein crystallography or cryo-electron microscopy (Desmyter et al. 2015). This is why the eminent structural biologist Karl-Peter Hopfner (LMU) turned to ChromoTek GmbH to produce nanobodies for a particularly tricky target, the giant protein complex INO80.
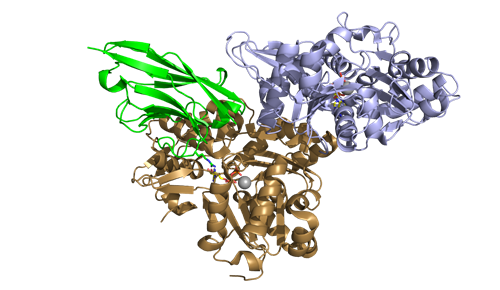
Figure 1: The crystal structure of the complex of ChromoTek's Nanobody VHH NactNB (green) with the IN080 subunits Arp4 (blue), nuclear actin (N-actin, brown). ATP molecules are shown as sticks. PDB code 5nbm; see Knoll et al. 2018 for more images.
The INO80 complex is a chromatin remodeller found in eukaryotic organisms from yeast to humans. Residing in the cell nucleus, INO80 takes care of the correct positioning of nucleosomes and their histone composition and is thus a cornerstone in the processes of gene expression and DNA replication. The whole INO80 complex comprises 15 proteins with a total mass of 1.3 MDa. It is not surprising that such an important protein complex is studied by many labs, but has often frustrated any attempt to elucidate the atomic structure of this complex or its parts due to its size.
- ChromoTek GmbH has successfully developed nanobodies against the INO80 complex from yeast. One of these, named NactNB, can be used to immunoprecipitate the endogenous INO80 complex from yeast. Upon closer inspection, it turned out to bind the INO80 subunits Arp4 and nuclear actin (N-actin). These subunits had been known for some time to form a complex that can be purified by itself but was recalcitrant to crystallisation. Now, thanks to the Nanobody VHH NactNB, the Arp4/N-actin complex was finally crystallised and its structure solved (see Figure).
- The complex of the NactNB Nanobody VHH and Arp4/N-actin recapitulates the key benefits of using a VHH: the Nanobody VHH is small enough to bind into a cleft between Arp4 and N-actin. Residues of the complementarity-determining region 3 (CDR3) of the Nanobody VHH even stick right into the core of N-actin, the nucleotide-binding pocket. This binding mode leads to the stabilisation of the Arp4/N-actin complex, which enabled its crystallisation and also DNA-binding assays. Based on these and other results, the Hopfner lab can now propose a model how the INO80 complex processes nucleosomes.
ChromoTek’s Nanobody VHH NactNB thus joins an illustrious list of nanobodies that have helped the field of structural biology move forward. A significant number of high-impact protein structures have been solved with the aid of these small camelid antibody fragments (see e.g. Desmyter et al. 2015 for a review). Most prominent among these is arguably the Nanobody VHH dubbed “Nb80”, which helped to crystallise the active state of a G-protein coupled receptor (GPCR, Rasmussen et al. 2011) - and thus led to a Nobel prize for Brian Kobilka (2012).
Knoll KR, Eustermann S, Niebauer V, Oberbeckmann E, Stoehr G, Schall K, Tosi A, Schwarz M, Buchfellner A, Korber P, Hopfner KP. The nuclear actin-containing Arp8 module is a linker DNA sensor driving INO80 chromatin remodeling. Nature Structural & Molecular Biology 2018, 25(9):823-832.
Desmyter A, Spinelli S, Roussel A, Cambillau C. Camelid nanobodies: killing two birds with one stone. Current Opinion in Structural Biology 2015, 32:1-8
Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, Devree BT, Rosenbaum DM, Thian FS, Kobilka TS, Schnapp A, Konetzki I, Sunahara RK, Gellman SH, Pautsch A, Steyaert J, Weis WI, Kobilka BK. Structure of a nanobody-stabilized active state of the β(2) adrenoceptor. Nature 2011, 469(7329):175-80
