Introduction to Immunoprecipitation
An overview of the immunoprecipitation (IP) application.
What is Immunoprecipitation?
Immunoprecipitation (IP) is a precipitation technique that purifies and enriches a protein of interest, allowing the identification of protein-protein interactions in proteomics workflows. IP is an important technique used to investigate the presence, relative abundance, size, upregulation or downregulation, stability, post-translational modifications (PTMs), and interactions between proteins.
Related: 8 tips on how to perform better immunoprecipitation experiments
IP is used to isolate a single protein (individual protein immunoprecipitation, the target antigen of the antibody). Variations of IP (e.g., Co-Immunoprecipitation, Co-IP) are used to study the interactions between the primary antigen protein and other proteins or nucleic acids (e.g., Chromatin Immunoprecipitation, ChIP, or RNA Immunoprecipitation, RIP) (Figure 1).
Learn more about the different types of immunoprecipitation below:
Individual Protein Immunoprecipitation (IP)Individual Protein Immunoprecipitation uses an antibody to isolate a selected protein of interest from cell lysate. The antibody binds to the protein and the antibody/antigen complex is pulled out of the sample using protein A/G-coupled agarose or magnetic beads. The beads are washed and the protein is eluted. Purified antigen obtained by IP is verified by a variety of molecular techniques (e.g., ELISA and Western blot), and isolated proteins are quantified and identified by mass spectrometry using enzymatic digestion patterns based on the primary sequence (Figure 1). |
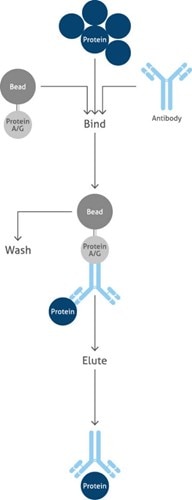 |
Overview of the main steps of Individual Protein Immunoprecipitation (IP) |
Co-Immunoprecipitation (Co-IP)Co-Immunoprecipitation (Co-IP) is a powerful tool used to analyze protein–protein interactions. The main purpose of Co-IP is the identification of interaction partners (other proteins, ligands, co-factors, or signaling molecules) to the protein of interest. It is an effective process used to separate proteins from serum, cell lysate, homogenized tissue, or conditioned media (Figure 2). |
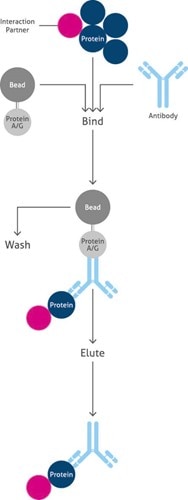 |
Overview of the main steps of Co-Immunoprecipitation (Co-IP) |
Chromatin Immunoprecipitation (ChIP)Chromatin Immunoprecipitation (ChIP) is a type of immunoprecipitation used to investigate regions of the genome associated with a target DNA-binding protein, or conversely to identify specific proteins associated with a particular region of the genome. It is commonly used in epigenetics research (e.g., ChIP monitors transcriptional regulation via modifications of histones) (Figure 3). For the full Proteintech ChIP protocol, see ChIP protocol section. |
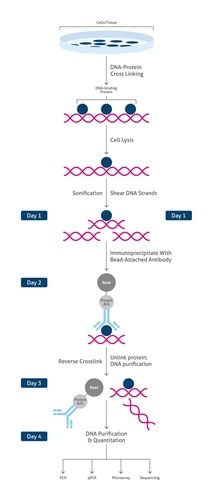 |
Overview of the main steps of Chromatin Immunoprecipitation (ChIP) |
RNA Immunoprecipitation (RIP)RNA Immunoprecipitation (RIP) targets RNA-binding proteins (ribonucleoproteins). It is performed using an antibody that targets a specific RNA-binding protein. The RNA-protein complexes are separated by RNA extraction. Some variants of RIP, such as photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) include cross-linking steps, which then require less careful lysis conditions (Figure 4). |
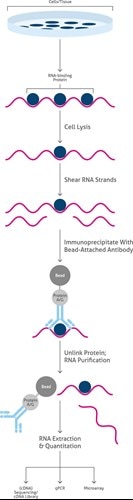 |
Overview of the main steps of RNA Immunoprecipitation (RIP). |