Tested Applications
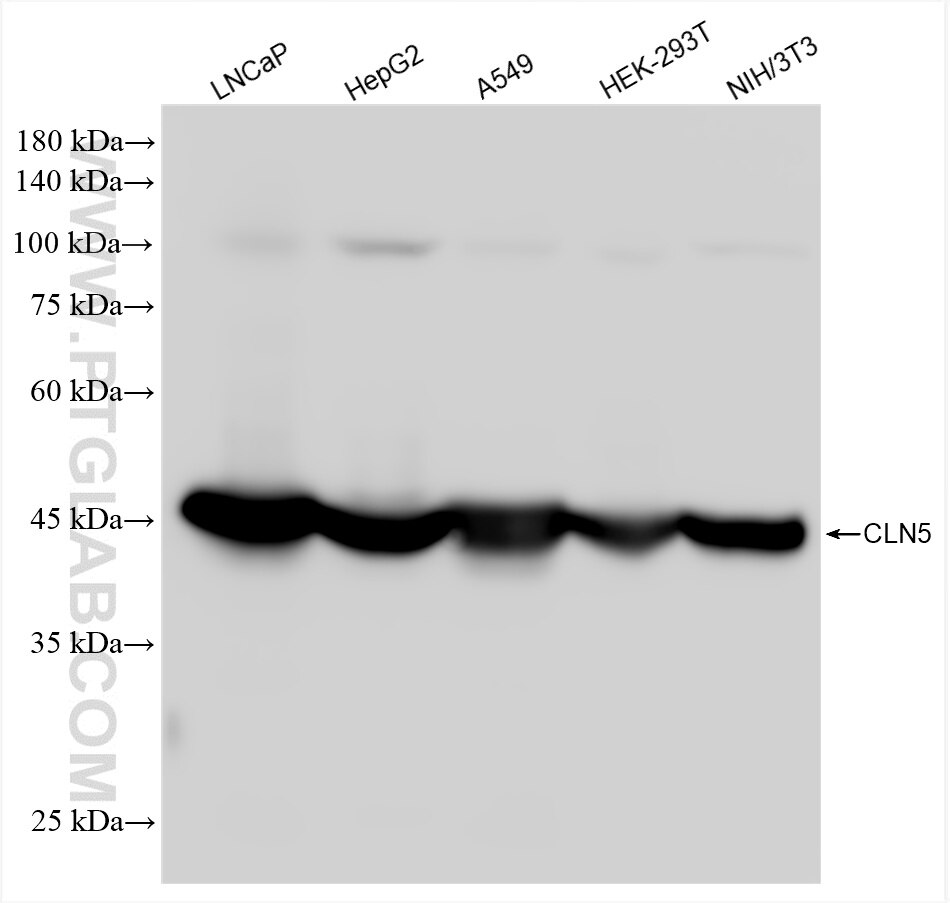
| Positive WB detected in | LNCaP cells, HepG2 cells, A549 cells, HEK-293T cells, NIH/3T3 cells |
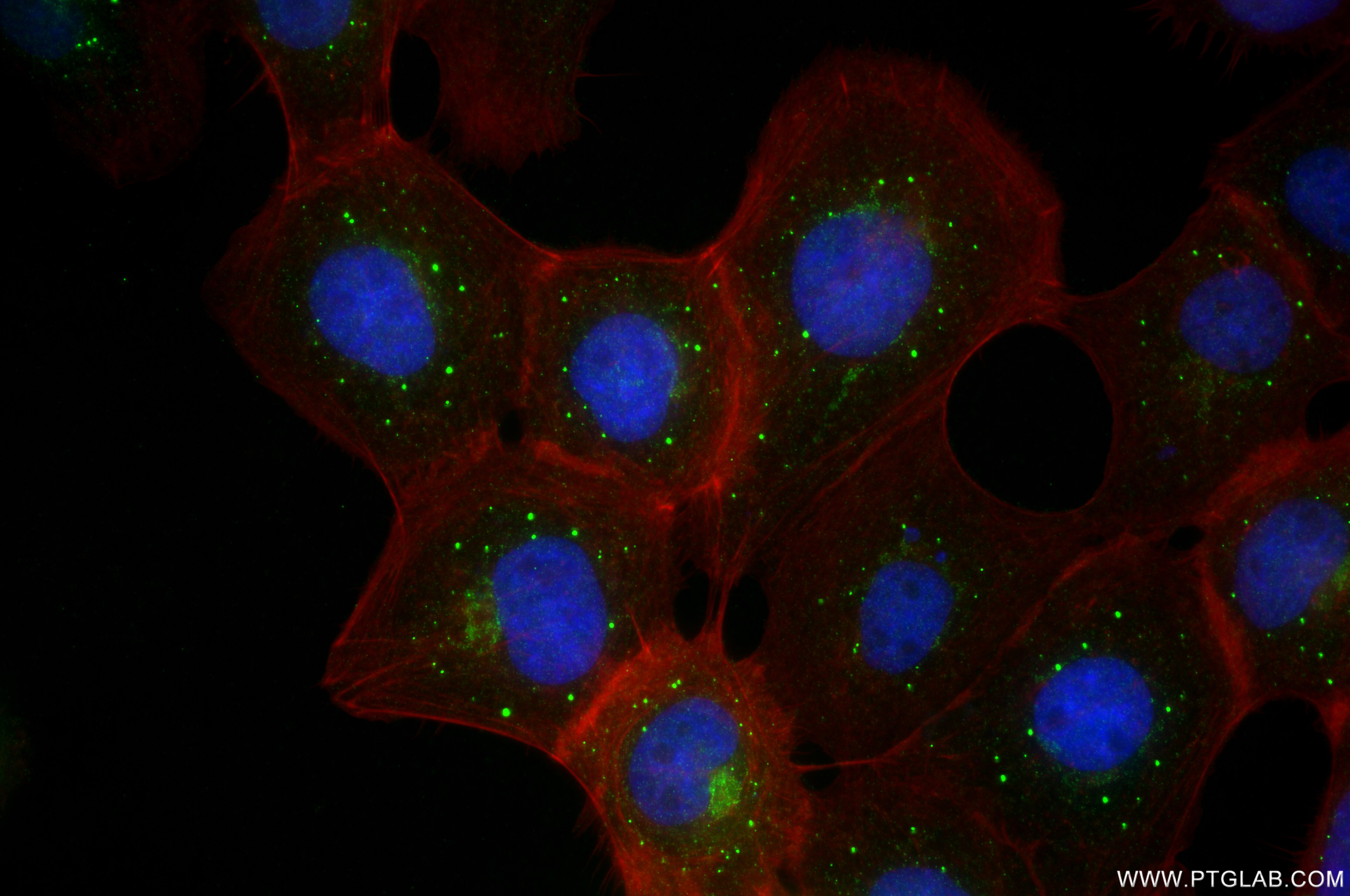
| Positive IF/ICC detected in | A431 cells |
Recommended dilution
| Application | Dilution |
|---|---|
| Western Blot (WB) | WB : 1:5000-1:50000 |
| Immunofluorescence (IF)/ICC | IF/ICC : 1:250-1:1000 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
85378-1-RR targets CLN5 in WB, IF/ICC, ELISA applications and shows reactivity with human, mouse samples.
| Tested Reactivity | human, mouse |
| Host / Isotype | Rabbit / IgG |
| Class | Recombinant |
| Type | Antibody |
| Immunogen |
CatNo: Ag18175 Product name: Recombinant human CLN5 protein Source: e coli.-derived, PGEX-4T Tag: GST Domain: 107-407 aa of BC153154 Sequence: KRFDFRPKPDPYCQAKYTFCPTGSPIPVMEGDDDIEVFRLQAPVWEFKYGDLLGHLKIMHDAIGFRSTLTGKNYTMEWYELFQLGNCTFPHLRPEMDAPFWCNQGAACFFEGIDDVHWKENGTLVQVATISGNMFNQMAKWVKQDNETGIYYETWNVKASPEKGAETWFDSYDCSKFVLRTFNKLAEFGAEFKNIETNYTRIFLYSGEPTYLGNETSVFGPTGNKTLGLAIKRFYYPFKPHLPTKEFLLSLLQIFDAVIVHKQFYLFYNFEYWFLPMKFPFIKITYEEIPLPIRNKTLSGL Predict reactive species |
| Full Name | ceroid-lipofuscinosis, neuronal 5 |
| Calculated Molecular Weight | 407 aa, 46 kDa |
| Observed Molecular Weight | 48 kDa |
| GenBank Accession Number | BC153154 |
| Gene Symbol | CLN5 |
| Gene ID (NCBI) | 1203 |
| Conjugate | Unconjugated |
| Form | Liquid |
| Purification Method | Protein A purification |
| UNIPROT ID | O75503 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol, pH 7.3. |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. 20ul sizes contain 0.1% BSA. |
Background Information
The CLN5 (ceroid lipofuscinosis neuronal protein 5) protein is ubiquitously expressed in the majority of tissues studied and in the brain, CLN5 shows both neuronal and glial cell expression. CLN5 plays a role in maintaining an acidic environment in the lysosomes, a critical feature for a functional lysosome (PMID: 38236309). Moreover, CLN5-deficient neuronal progenitor cells showed reduced thioesterase activity, confirming the live cell function of CLN5 in setting S-depalmitoylation levels (PMID: 35427157).
Protocols
| Product Specific Protocols | |
|---|---|
| IF protocol for CLN5 antibody 85378-1-RR | Download protocol |
| WB protocol for CLN5 antibody 85378-1-RR | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |