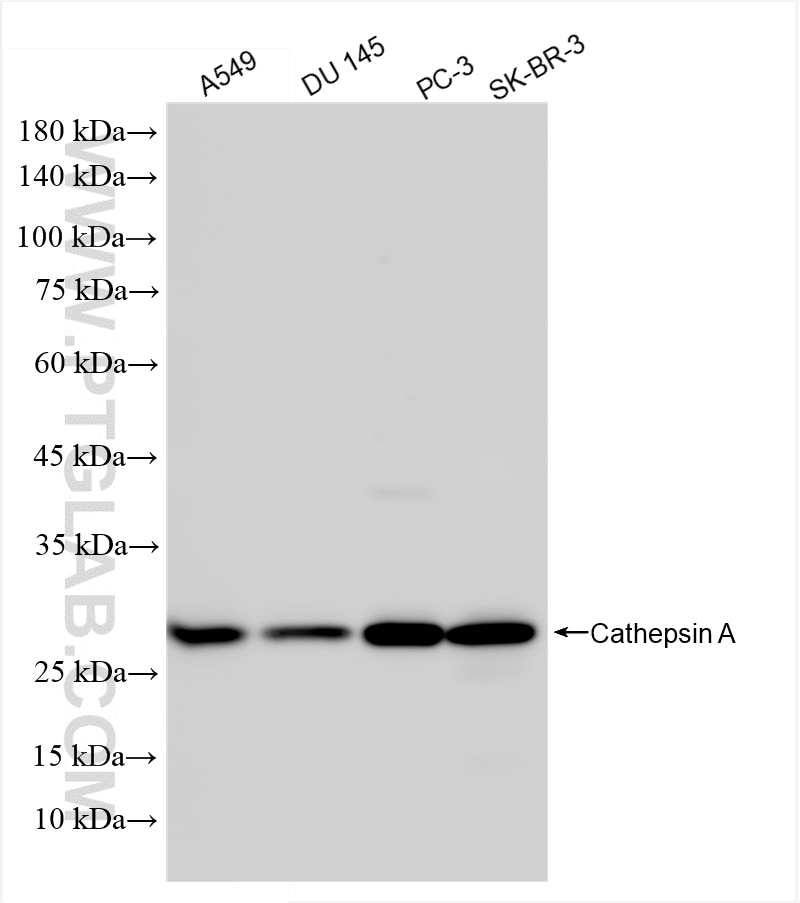
Tested Applications
| Positive WB detected in | A549 cells, DU 145 cells, PC-3 cells, SK-BR-3 cells |
Recommended dilution
| Application | Dilution |
|---|---|
| Western Blot (WB) | WB : 1:1000-1:6000 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
80272-3-RR targets CTSA in WB, ELISA applications and shows reactivity with human samples.
| Tested Reactivity | human |
| Host / Isotype | Rabbit / IgG |
| Class | Recombinant |
| Type | Antibody |
| Immunogen | CTSA fusion protein Ag7073 Predict reactive species |
| Full Name | cathepsin A |
| Calculated Molecular Weight | 54 kDa |
| Observed Molecular Weight | 54-60, 32, 20 kDa |
| GenBank Accession Number | BC000597 |
| Gene Symbol | Cathepsin A |
| Gene ID (NCBI) | 5476 |
| Conjugate | Unconjugated |
| Form | Liquid |
| Purification Method | Protein A purification |
| UNIPROT ID | P10619 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol, pH 7.3. |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. 20ul sizes contain 0.1% BSA. |
Background Information
CTSA is also named as PPGB(protective protein for beta-galactosidase), PPCA(protective protein cathepsin A), cathepsin A, carboxypeptidase C, carboxypeptidase L and belongs to the peptidase S10 family. It is a ubiquitously expressed multifunctional enzyme, with deamidase, esterase, and carboxypeptidase activities and a preference for substrates with hydrophobic amino acid residues at the P1-prime position. It can be cleaved into the following 2 chains and defects in CTSA are the cause of galactosialidosis (GSL). This protein has 2 glycosylation sites. CTSA is synthesized as a 54-kDa precursor protein and composed of 32- and 20-kDa subunits linked together by disulfide bonds(PMID: 16461364,19574551).
Protocols
| Product Specific Protocols | |
|---|---|
| WB protocol for CTSA antibody 80272-3-RR | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |