Tested Applications
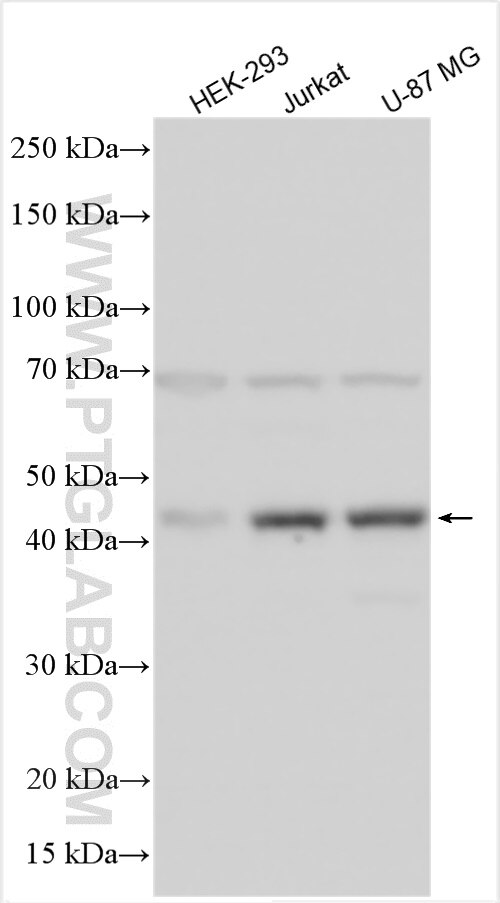
| Positive WB detected in | HEK-293 cells |
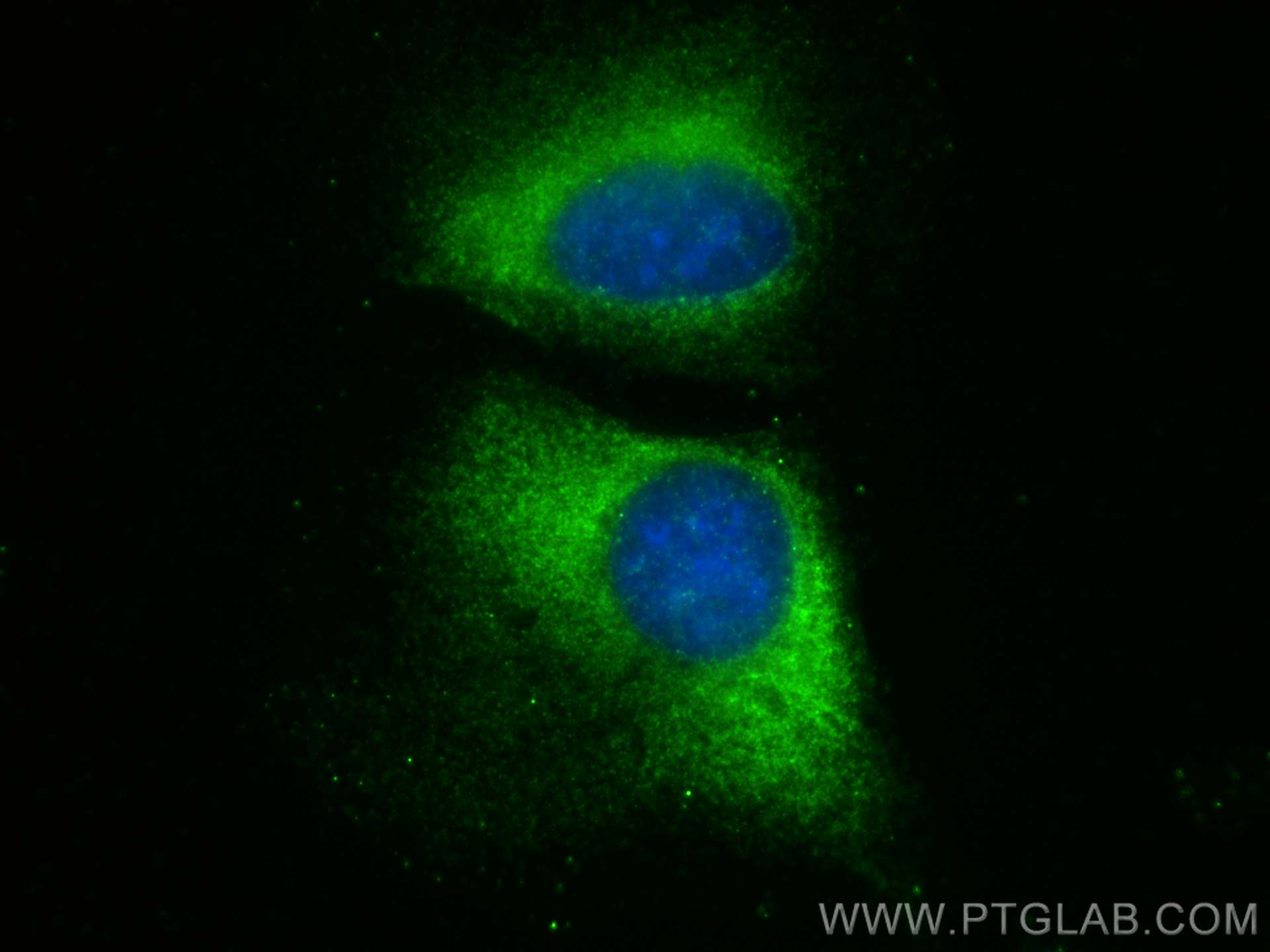
| Positive IF/ICC detected in | U2OS cells |
Recommended dilution
| Application | Dilution |
|---|---|
| Western Blot (WB) | WB : 1:1000-1:6000 |
| Immunofluorescence (IF)/ICC | IF/ICC : 1:200-1:800 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
25149-1-AP targets NAT12 in WB, IF/ICC, ELISA applications and shows reactivity with human samples.
| Tested Reactivity | human |
| Host / Isotype | Rabbit / IgG |
| Class | Polyclonal |
| Type | Antibody |
| Immunogen |
CatNo: Ag18435 Product name: Recombinant human NAT12 protein Source: e coli.-derived, PGEX-4T Tag: GST Domain: 1-362 aa of BC118589 Sequence: MAEVPPGPSSLLPPPAPPAPAAVEPRCPFPAGAALACCSEDEEDDEEHEGGGSRSPAGGESATVAAKGHPCLRCPQPPQEQQQLNGLISPELRHLRAAASLKSKVLSVAEVAATTATPDGGPRATATKGAGVHSGERPPHSLSSNARTAVPSPVEAAAASDPAAARNGLAEGTEQEEEEEDEQVRLLSSSLTADCSLRSPSGREVEPGEDRTIRYVRYESELQMPDIMRLITKDLSEPYSIYTYRYFIHNWPQLCFLAMVGEECVGAIVCKLDMHKKMFRRGYIAMLAVDSKYRRNGIGTNLVKKAIYAMVEGDCDEVVLETEITNKSALKLYENLGFVRDKRLFRYYLNGVDALRLKLWLR Predict reactive species |
| Full Name | N-acetyltransferase 12 (GCN5-related, putative) |
| Calculated Molecular Weight | 362 aa, 39 kDa |
| Observed Molecular Weight | 40 kDa |
| GenBank Accession Number | BC118589 |
| Gene Symbol | NAT12 |
| Gene ID (NCBI) | 122830 |
| RRID | AB_3085773 |
| Conjugate | Unconjugated |
| Form | Liquid |
| Purification Method | Antigen affinity purification |
| UNIPROT ID | Q147X3 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol, pH 7.3. |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. 20ul sizes contain 0.1% BSA. |
Background Information
N-terminal acetylation (Nt-acetylation) by N-terminal acetyltransferases (NATs) is one of the most common protein modifications in eukaryotes. The NatC complex represents one of three major NATs of which the substrate profile remains largely unexplored. NAT12/NAA30 (N-alpha-acetyltransferase 30) is the catalytic subunit of the NAT-C complex (PMID: 27694331, 26292663). The expected sizes of the two isoforms of the protein were 35 and 39 kDa. Western blots detected two bands of NAA12, the expected ~40 kDa band and a possibly dimer at ~70 kDa.
Protocols
| Product Specific Protocols | |
|---|---|
| IF protocol for NAT12 antibody 25149-1-AP | Download protocol |
| WB protocol for NAT12 antibody 25149-1-AP | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |