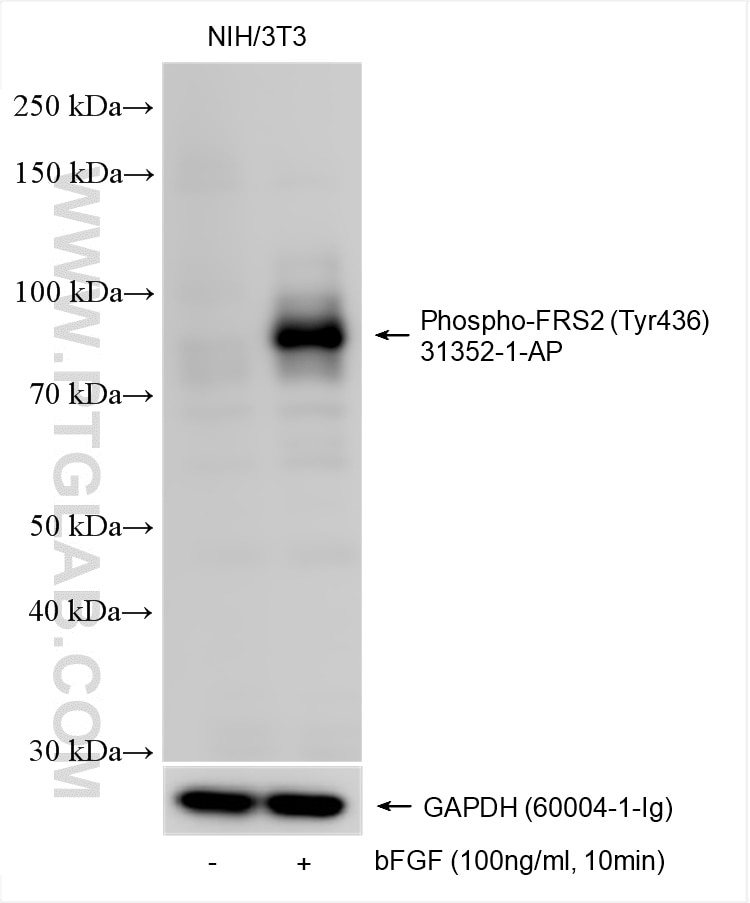
Tested Applications
| Positive WB detected in | bFGF treated NIH/3T3 cells |
Recommended dilution
| Application | Dilution |
|---|---|
| Western Blot (WB) | WB : 1:500-1:2000 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
31352-1-AP targets Phospho-FRS2 (Tyr436) in WB, ELISA applications and shows reactivity with human, mouse samples.
| Tested Reactivity | human, mouse |
| Host / Isotype | Rabbit / IgG |
| Class | Polyclonal |
| Type | Antibody |
| Immunogen |
Peptide Predict reactive species |
| Full Name | fibroblast growth factor receptor substrate 2 |
| Calculated Molecular Weight | 60 kDa |
| Observed Molecular Weight | 80-85 kDa |
| GenBank Accession Number | BC021562 |
| Gene Symbol | FRS2 |
| Gene ID (NCBI) | 10818 |
| RRID | AB_3669956 |
| Conjugate | Unconjugated |
| Form | Liquid |
| Purification Method | Antigen affinity purification |
| UNIPROT ID | Q8WU20 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol, pH 7.3. |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. 20ul sizes contain 0.1% BSA. |
Background Information
Fibroblast growth factor substrate 2 (FRS2), a lipid-anchored docking protein that is phosphorylated upon activation of FGFR, is critical for recruitment of downstream signaling molecules and links the FGFRs to the Ras/Mek/Erk pathway and the PI3-Kinase/Akt pathway. Studies have found that the phosphotyrosine binding domain of FRS-2 directly binds the Trk receptors at the same phosphotyrosine residue that binds the signaling adapter Shc, suggesting a model in which competitive binding between FRS-2 and Shc regulates differentiation versusproliferation. (PMID: 19053057, PMID: 10092678)
Protocols
| Product Specific Protocols | |
|---|---|
| WB protocol for Phospho-FRS2 (Tyr436) antibody 31352-1-AP | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |