ChromoTek's Nanobodies are versatile and proven affinity tools for multiplexed analysis of protein-protein interactions using Luminex® or Biacore™ assay technologies
Benefit from high binding affinities and extraordinary functional stabilities of Nanobodies
Benefit from high binding affinities and extraordinary functional stabilities of Nanobodies
Recently we have published an applications note that describes how to benefit from Nanobodies for the analysis of protein-protein interaction (PPI) analysis when used in bead-based protein assays (BPAs) in Luminex systems. This applications note is based on a recent publication of Ulrich Rothbauer’s lab at the NMI in Tübingen, Germany.
In addition Eduardo Della Pia and Karen Martinez from the Department of Chemistry & Nano-Science Center, University of Copenhagen, Copenhagen, Denmark have demonstrated how to benefit from Nanobodies for PPI analysis in Biacore Surface Plasmon Resonance systems. The application of ChromoTek’s anti-GFP Nanobodies is also described in the Biacore manual.
(Groll N. et al.)
Nanobodies used in Luminex system
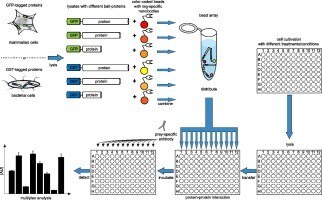
Groll N. et al. outline that knowledge on dynamic PPIs is essential to understand cellular processes and for the discovery and validation of compounds modulating such interactions. BPAs are emerging methodologies to analyze PPIs between bait- and prey-proteins. However, most studies still employ bacterial derived bait-proteins, which are of limited use since they lack posttranslational modifications or do not undergo correct folding upon heterologous expression. This application note reports on a novel approach to generate BPAs combining μ-scale purification of bait proteins combined with site-directed immobilization. Bait-proteins are produced as GST- or GFP-fusion constructs in bacterial or mammalian cells and one-step purified and immobilized from crude lysates using ChromoTek’s high affinity tag-specific nanobodies: GFP-Trap® or GST-Trap coupled to color-coded beads. Finally, those bait-coupled beads are combined in a protein-array for miniaturized multiplexed GST- and GFP pulldown studies. Recently this technology was successfully applied to study dynamic changes of the interaction between the endogenous prey-protein β-catenin and a set of individual bait proteins following proteasomal inhibition or signaling pathway perturbation.
Finally Groll N. et al. concluded that the application of high affinity tag-specific nanobodies enables an efficient, fast and reproducible generation of bead-based protein arrays directly from minute amounts of eukaryotic and prokaryotic protein expression cultures. They demonstrated the advantages of eukaryotic expression systems by the analysis of the binding behavior of the signaling protein β-catenin with three of its interaction partners. Furthermore it is proposed that this approach is suitable to study the effect of small molecules on proteins in cellular pathways by BPAs in a medium- to high-throughput mode. Identification of novel high affinity tag-nanobodies specific for additional protein or peptide-tags will expand the opportunities of such an approach substantially.
Nanobodies used in Biacore system
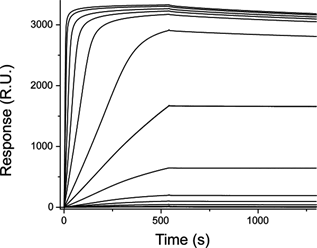
In this study Della Pia and Martinez demonstrated the potential of using single domain antibodies as capturing molecules in biosensing applications. Anti-green fluorescent protein Nanobodies were bound onto biosensor surfaces by using a variety of immobilization strategies. The interaction with the specific target of the Nanobody was characterized by surface plasmon resonance. Immobilized Nanobodies show high affinities for their antigens with KD = 3–6nM and outperform other antibody partners as capturing molecules facilitating also the data analysis. Furthermore they offer high resistance and stability to a wide range of denaturing agents. These unique biophysical properties and the production of novel single domain antibodies against affinity tags make them particularly attractive for use in biosensing and diagnostic assays.
Nanobodies are single domain antibodies are recombinantly expressed functional antibodies devoid of light chains. These binding elements are derived from heavy chain antibodies found in camelids and offer several distinctive properties. Nanobodies are of particular benefit for automated PPI analysis, because
- Nanobodies are small size binders of just 15 kDa, therefore more binder can be coupled to the surface area of a bead or slide. This results in an increased dynamic range and higher signal density and thus sensitivity of your protein-protein interaction study.
- Nanobodies have low nano Molar to pico Molar dissociation constants KD, which results in ultra-strong and fast binding of the protein of interest. This makes the experiment robust and quick (short incubation times)
- Nanobodies are stable and bind their antigens even under extreme conditions including chaotropic reagents and elevated temperatures. This enables you to apply harsh wash conditions to remove unspecific bonding and therefore reduces the background resulting in an improved assay sensitivity.
- Nanobodies are recombinant proteins and ChromoTek produces Nanobodies at high quality in a very reproducible manner without lot to lot variations making even large assays and independent from the production quality of the binder.
The following ChromoTek Nanobodies are available to capture GFP-, RFP- and GST-fusion proteins:
- GFP VHH, recombinant binding protein
- RFP VHH, recombinant binding protein
- GST VHH, recombinant binding protein
Interested in sample? Please send an email to support@chromotek.com.