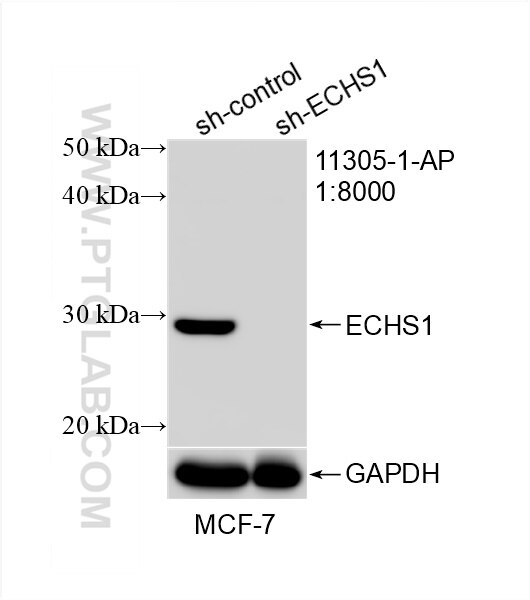
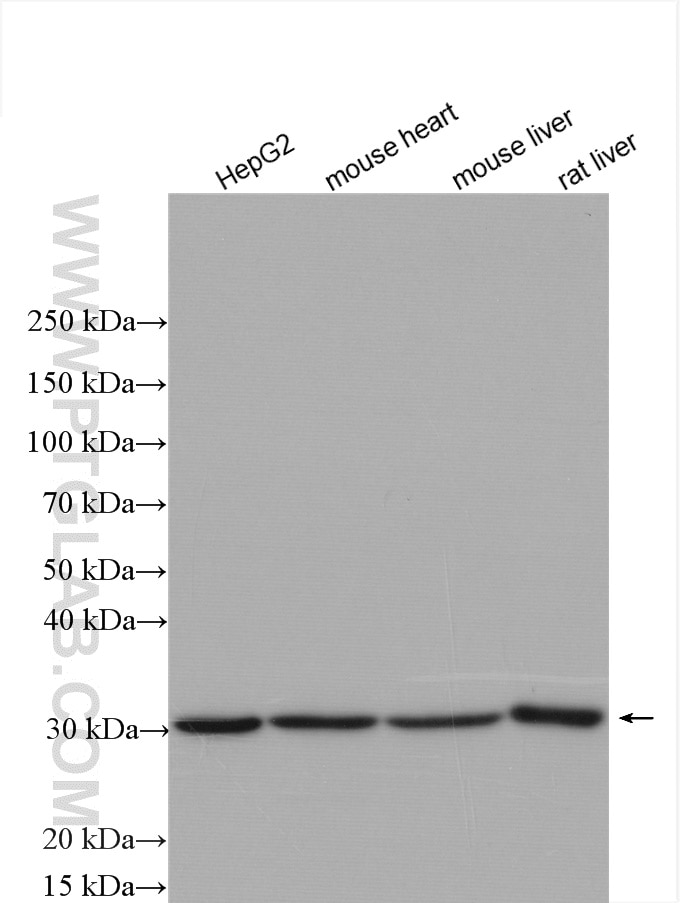
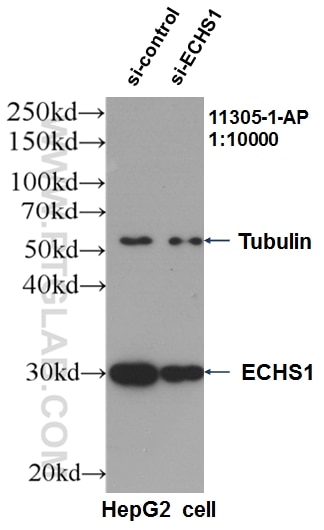
Tested Applications
| Positive WB detected in | HepG2 cells, HeLa cells, MCF-7 cells, mouse heart tissue, mouse liver tissue, rat liver tissue, L02 cells, rat heart tiisue |
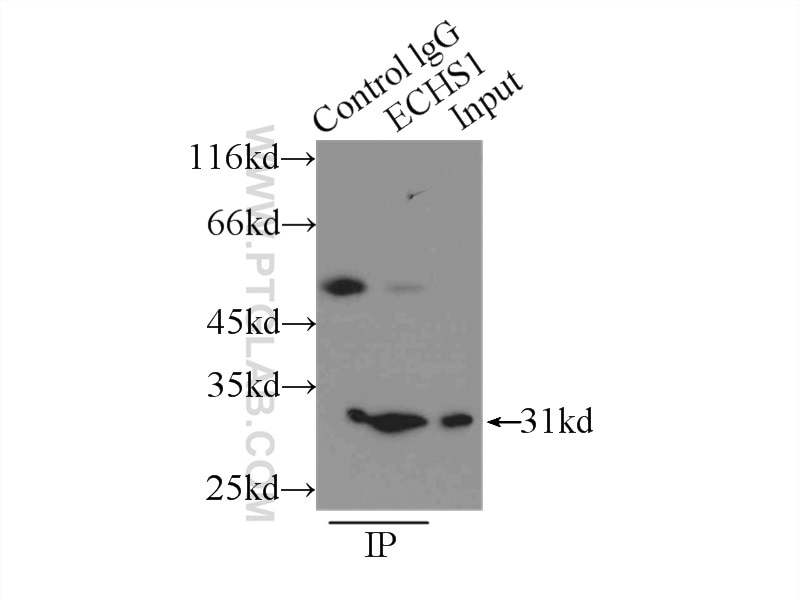
| Positive IP detected in | PC-3 cells |
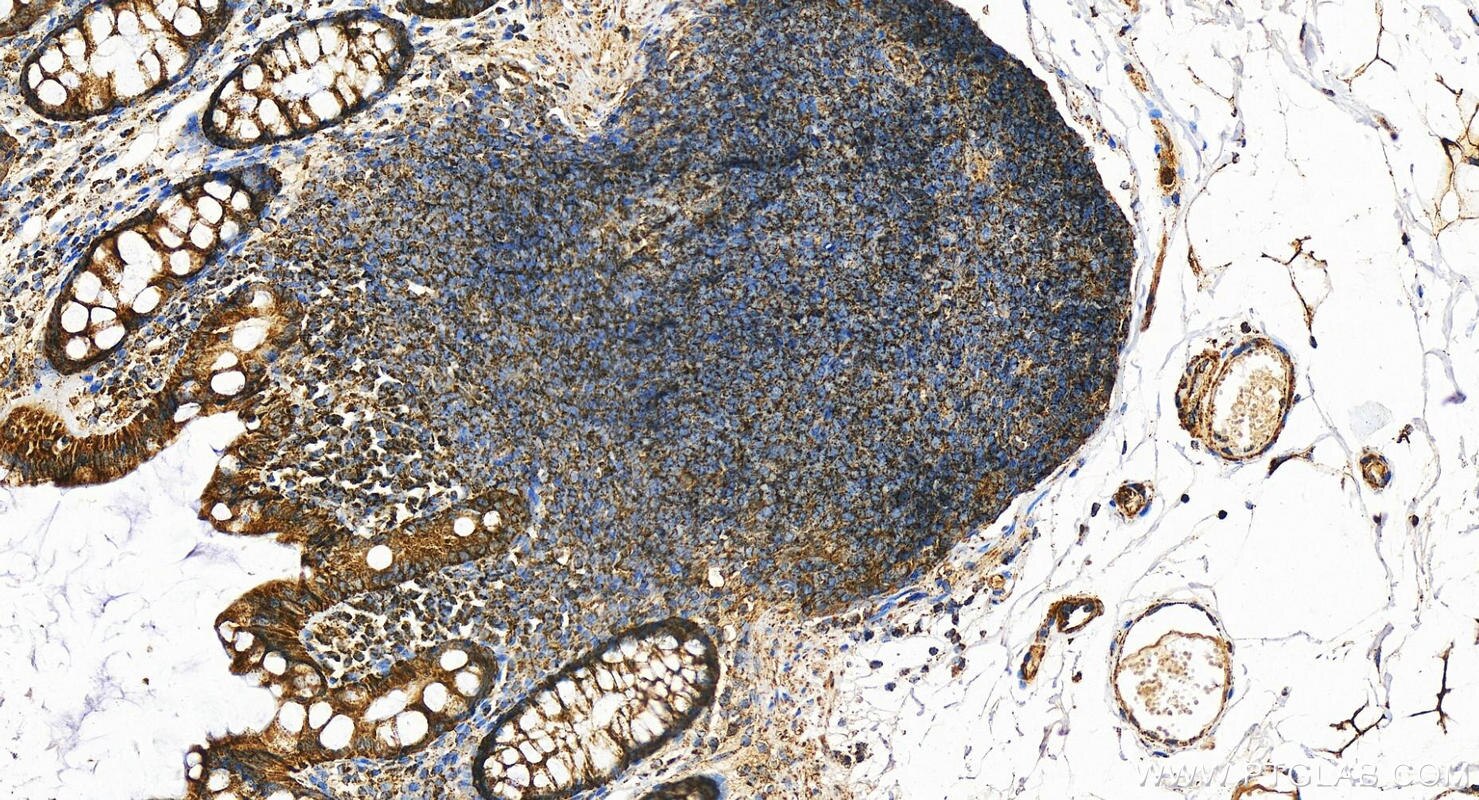
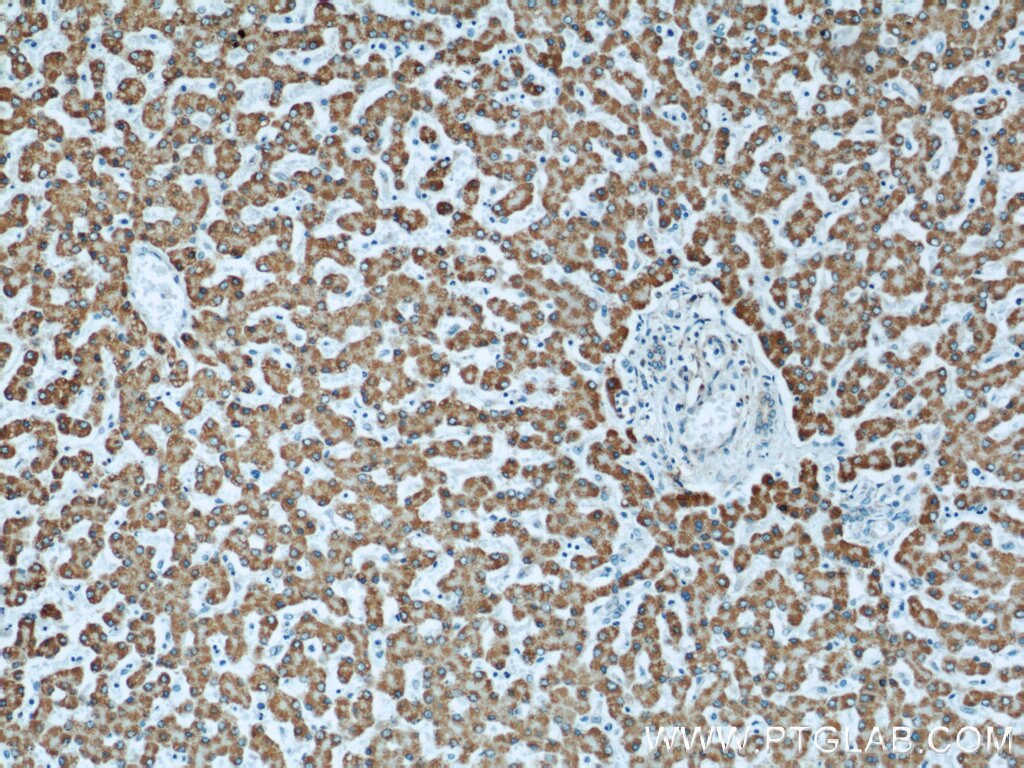
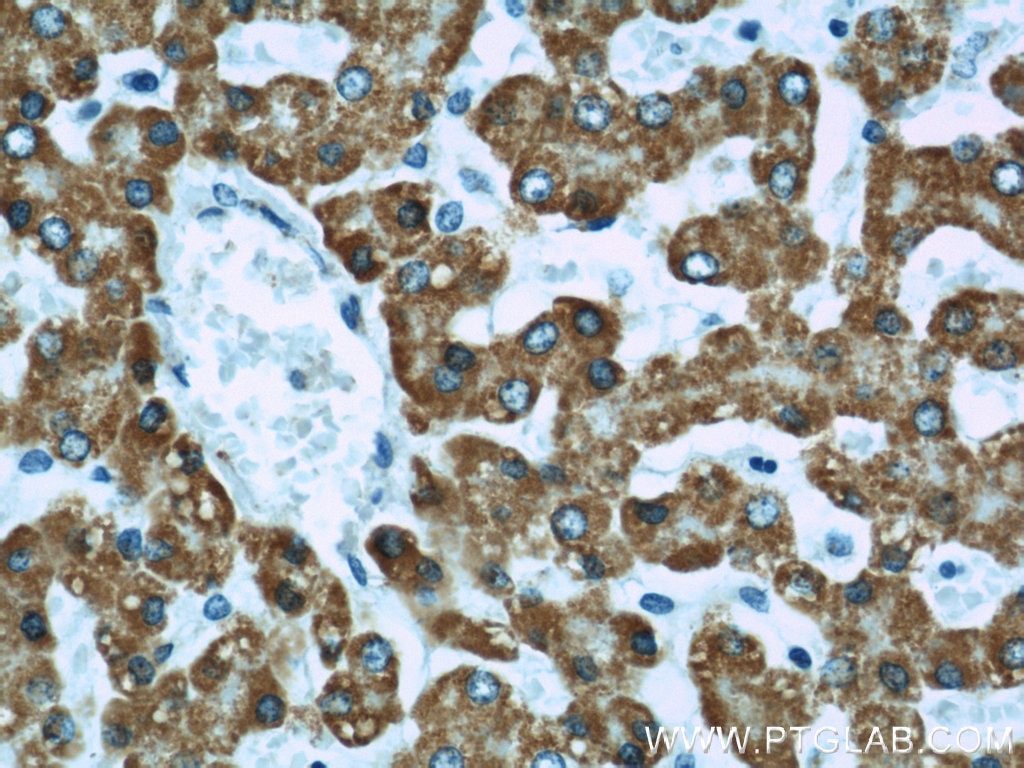
| Positive IHC detected in | human colon tissue, human liver tissue Note: suggested antigen retrieval with TE buffer pH 9.0; (*) Alternatively, antigen retrieval may be performed with citrate buffer pH 6.0 |
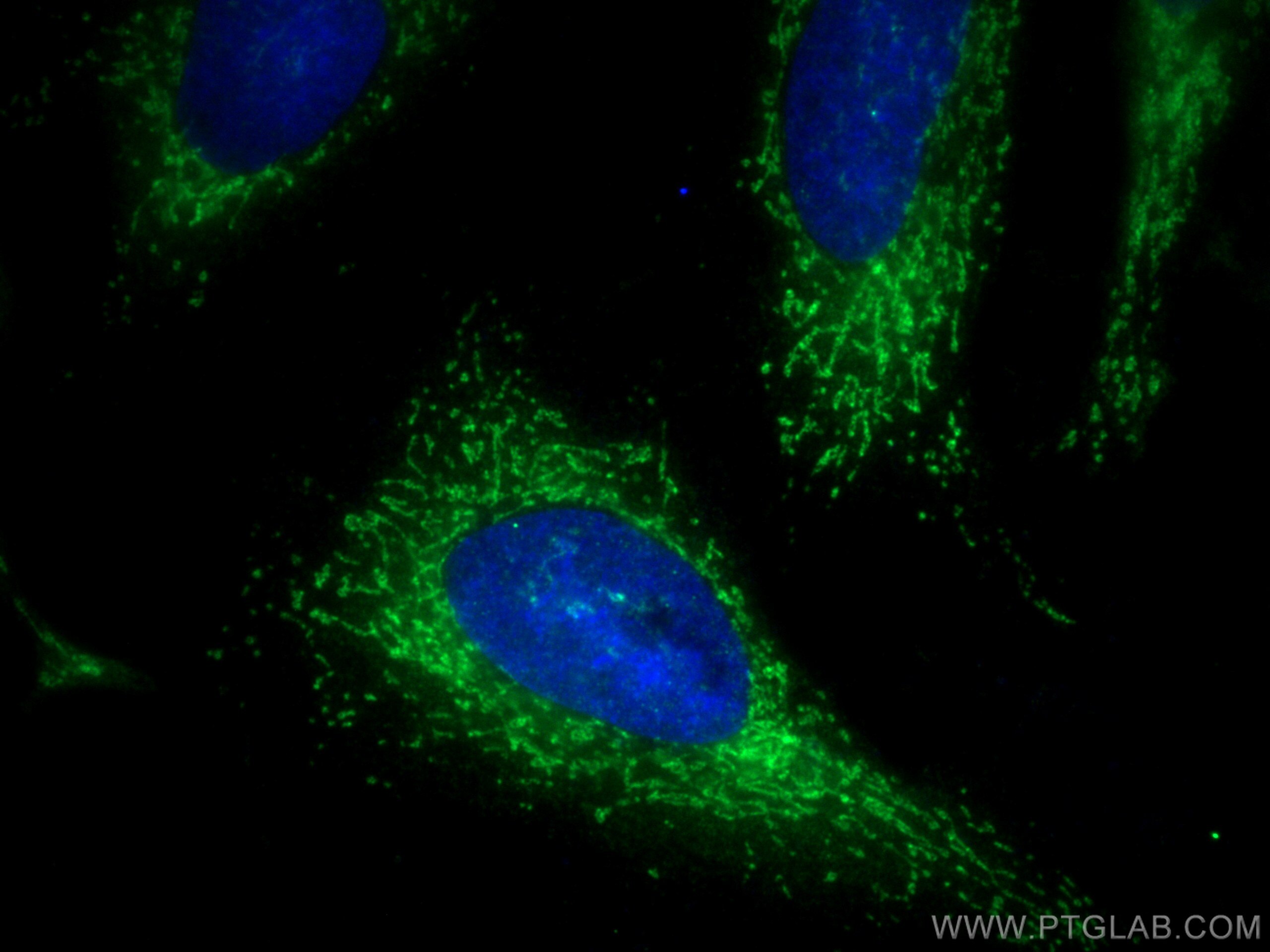
| Positive IF/ICC detected in | HeLa cells |
Recommended dilution
| Application | Dilution |
|---|---|
| Western Blot (WB) | WB : 1:5000-1:50000 |
| Immunoprecipitation (IP) | IP : 0.5-4.0 ug for 1.0-3.0 mg of total protein lysate |
| Immunohistochemistry (IHC) | IHC : 1:600-1:2400 |
| Immunofluorescence (IF)/ICC | IF/ICC : 1:200-1:800 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Published Applications
| KD/KO | See 2 publications below |
| WB | See 29 publications below |
| IHC | See 3 publications below |
| IF | See 2 publications below |
| IP | See 1 publications below |
Product Information
11305-1-AP targets ECHS1 in WB, IHC, IF/ICC, IP, ELISA applications and shows reactivity with human, mouse, rat samples.
| Tested Reactivity | human, mouse, rat |
| Cited Reactivity | human, mouse, rat |
| Host / Isotype | Rabbit / IgG |
| Class | Polyclonal |
| Type | Antibody |
| Immunogen |
CatNo: Ag1842 Product name: Recombinant human ECHS1 protein Source: e coli.-derived, PGEX-4T Tag: GST Domain: 1-290 aa of BC008906 Sequence: MAALRVLLSCVRGPLRPPVRCPAWRPFASGANFEYIIAEKRGKNNTVGLIQLNRPKALNALCDGLIDELNQALKIFEEDPAVGAIVLTGGDKAFAAGADIKEMQNLSFQDCYSSKFLKHWDHLTQVKKPVIAAVNGYAFGGGCELAMMCDIIYAGEKAQFAQPEILIGTIPGAGGTQRLTRAVGKSLAMEMVLTGDRISAQDAKQAGLVSKICPVETLVEEAIQCAEKIASNSKIVVAMAKESVNAAFEMTLTEGSKLEKKLFYSTFATDDRKEGMTAFVEKRKANFKDQ Predict reactive species |
| Full Name | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial |
| Calculated Molecular Weight | 31 kDa |
| Observed Molecular Weight | 29-31 kDa |
| GenBank Accession Number | BC008906 |
| Gene Symbol | ECHS1 |
| Gene ID (NCBI) | 1892 |
| RRID | AB_2262166 |
| Conjugate | Unconjugated |
| Form | Liquid |
| Purification Method | Antigen affinity purification |
| UNIPROT ID | P30084 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol, pH 7.3. |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. 20ul sizes contain 0.1% BSA. |
Background Information
Enoyl-coenzyme A hydratase (ECHS1) is a mitochondrial protein which catalyzes the hydration of 2-trans-enoyl-coenzyme A intermediates to L-3-hydroxyacyl-coenzyme A, playing key role in metabolism of fatty acids in mitochondria. ECHS1 is highly expressed in muscle, liver and fibroblasts. Altered expression of ECHS1 has been considered as a characteristic feature of mitochondria dysfunction. (23275097, 23235493)
Protocols
| Product Specific Protocols | |
|---|---|
| IF protocol for ECHS1 antibody 11305-1-AP | Download protocol |
| IHC protocol for ECHS1 antibody 11305-1-AP | Download protocol |
| IP protocol for ECHS1 antibody 11305-1-AP | Download protocol |
| WB protocol for ECHS1 antibody 11305-1-AP | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |
Publications
| Species | Application | Title |
|---|---|---|
Drug Resist Updat MYC expression and fatty acid oxidation in EGFR-TKI acquired resistance | ||
Cancer Res One-hit effects in cancer: altered proteome of morphologically normal colon crypts in familial adenomatous polyposis. | ||
J Proteome Res Proteomic analysis of left ventricular remodeling in an experimental model of heart failure. | ||
J Proteome Res Novel Proteomic Biomarker Panel for Prediction of Aggressive Metastatic Hepatocellular Carcinoma Relapse in Surgically Resectable Patients. | ||
Oncotarget Quantitative proteomics reveals molecular mechanism of gamabufotalin and its potential inhibition on Hsp90 in lung cancer. | ||
Am J Pathol The Krebs cycle and mitochondrial mass are early victims of endothelial dysfunction: proteomic approach. |