Tested Applications
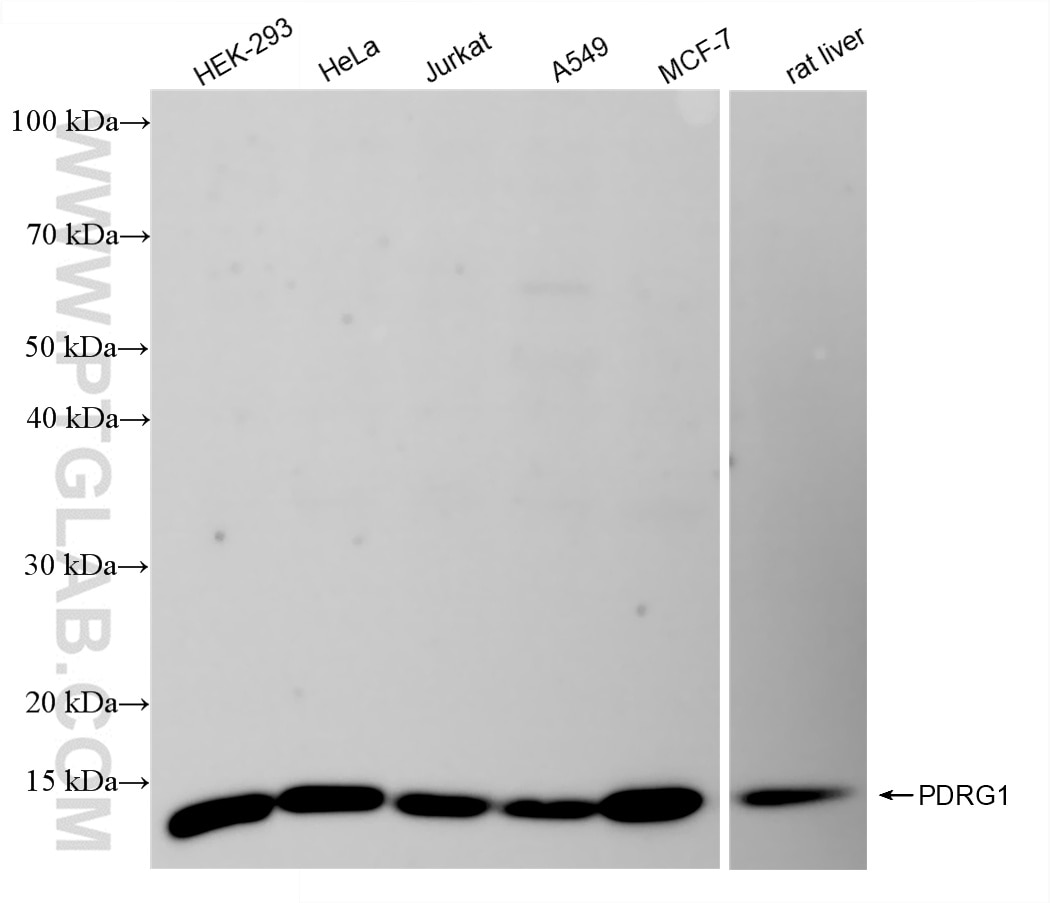
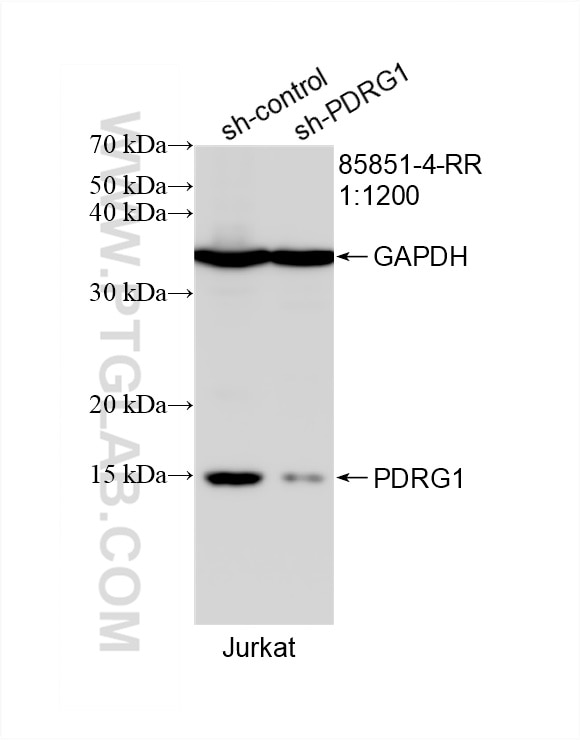
| Positive WB detected in | HEK-293 cells, Jurkat cells, HeLa cells, A549 cells, MCF-7 cells, rat liver tissue |
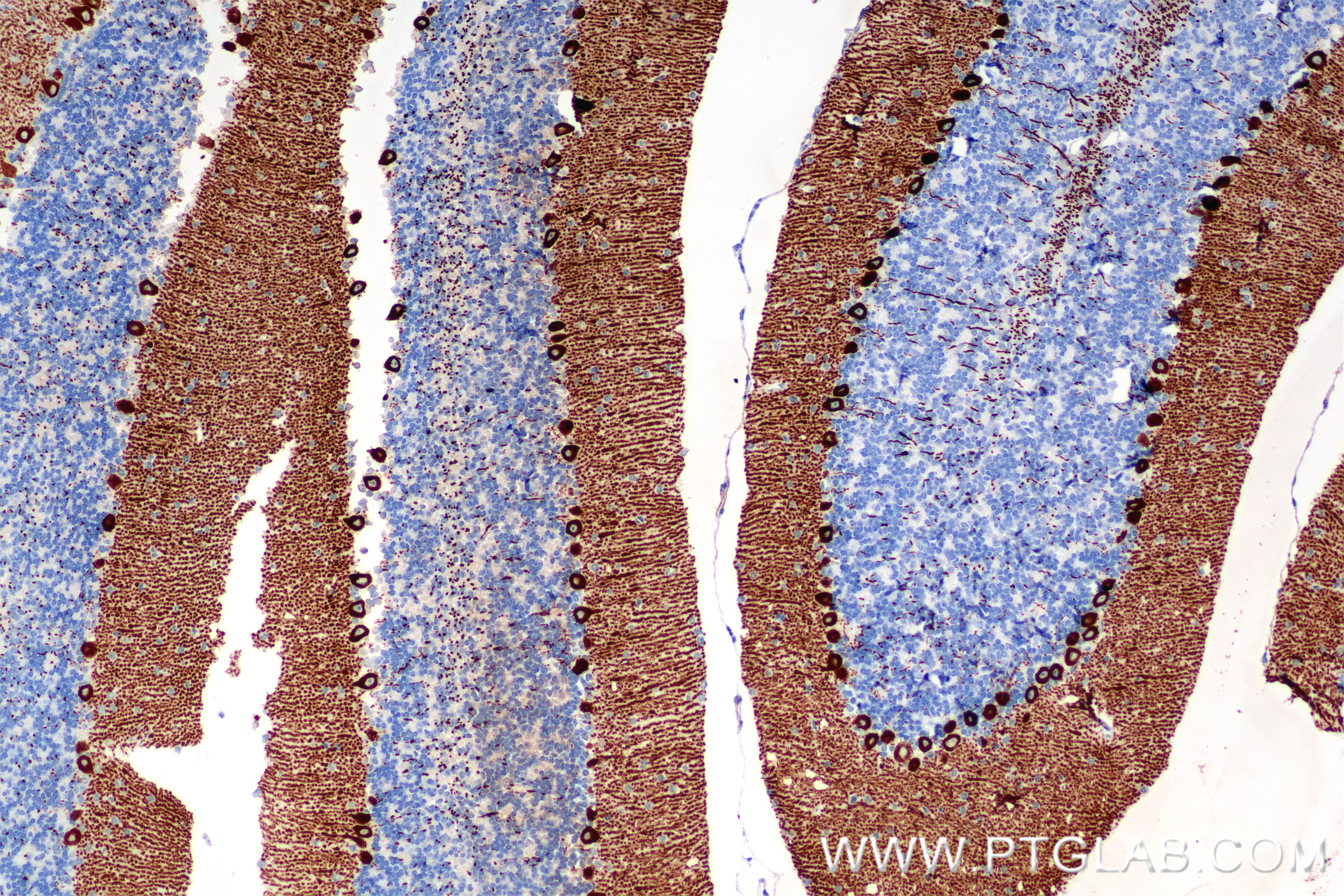
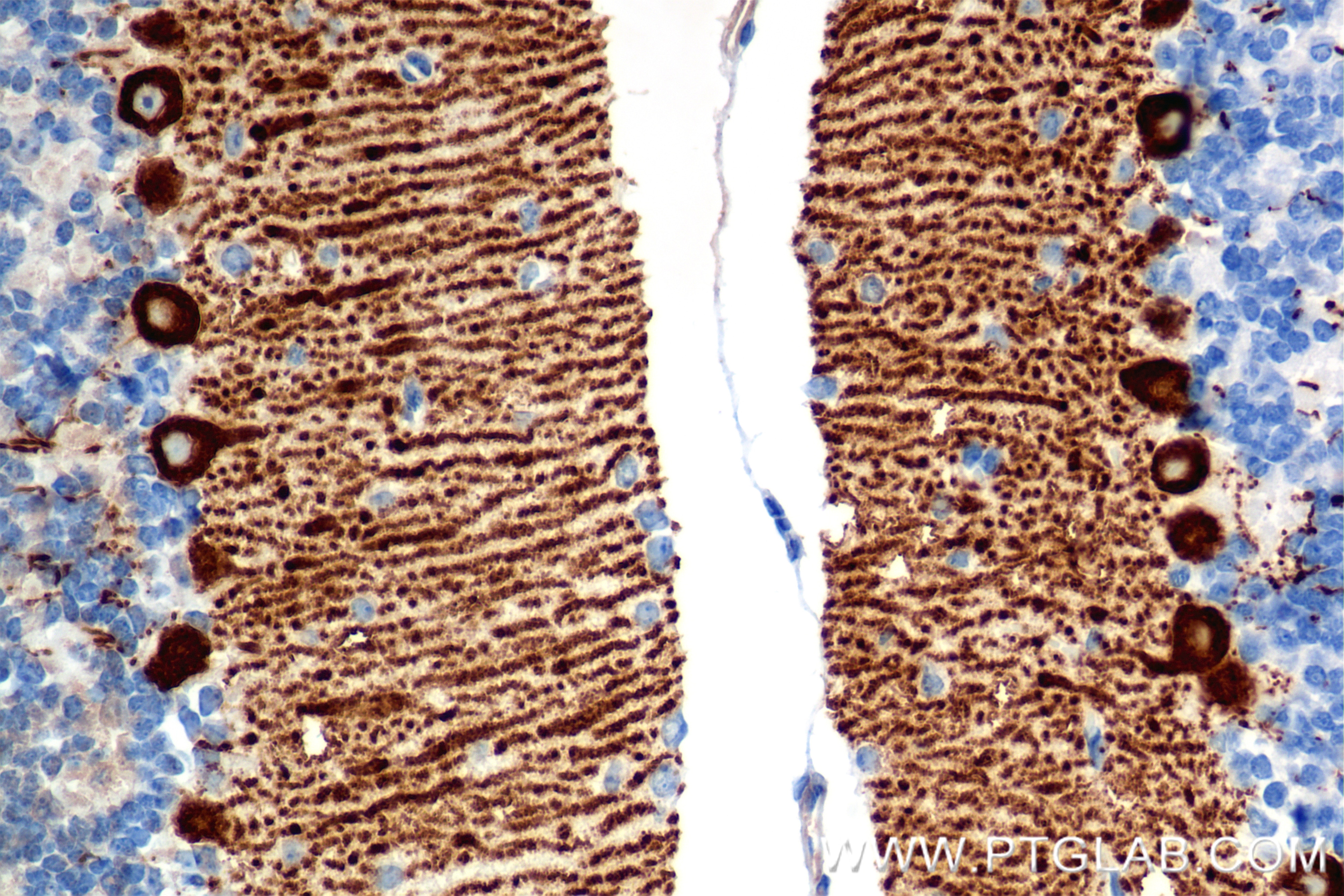
| Positive IHC detected in | mouse cerebellum tissue Note: suggested antigen retrieval with TE buffer pH 9.0; (*) Alternatively, antigen retrieval may be performed with citrate buffer pH 6.0 |
Recommended dilution
| Application | Dilution |
|---|---|
| Western Blot (WB) | WB : 1:5000-1:50000 |
| Immunohistochemistry (IHC) | IHC : 1:400-1:1600 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
85851-4-RR targets PDRG1 in WB, IHC, ELISA applications and shows reactivity with human, rat samples.
| Tested Reactivity | human, rat |
| Host / Isotype | Rabbit / IgG |
| Class | Recombinant |
| Type | Antibody |
| Immunogen |
CatNo: Ag9109 Product name: Recombinant human PDRG1 protein Source: e coli.-derived, PGEX-4T Tag: GST Domain: 1-133 aa of BC009334 Sequence: MLSPEAERVLRYLVEVEELAEEVLADKRQIVDLDTKRNQNREGLRALQKDLSLSEDVMVCFGNMFIKMPHPETKEMIEKDQDHLDKEIEKLRKQLKVKVNRLFEAQGKPELKGFNLNPLNQDELKALKVILKG Predict reactive species |
| Full Name | p53 and DNA damage regulated 1 |
| Calculated Molecular Weight | 16 kDa |
| Observed Molecular Weight | 15-16 kDa |
| GenBank Accession Number | BC009334 |
| Gene Symbol | PDRG1 |
| Gene ID (NCBI) | 81572 |
| Conjugate | Unconjugated |
| Form | Liquid |
| Purification Method | Protein A purification |
| UNIPROT ID | Q9NUG6 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol, pH 7.3. |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. 20ul sizes contain 0.1% BSA. |
Background Information
PDRG1 codes for a novel protein called p53 and DNA damage-regulated protein 1. Human PDRG1 is predominantly expressed in normal testis and its expression can be induced by UV irradiation and be repressed by p53 [PMID14562055]. Recently, PDRG1 was identified as a novel tumor marker for multiple malignancies that is selectively regulated by genotoxic stress. Its expression was upregulated in multiple malignancies including cancers of the colon, rectum, ovary, lung, stomach, breast and uterus when compared to normal tissues [PMID: 21193842], suggesting its being a novel tumor marker that could play a role in cancer development and/or progression.
Protocols
| Product Specific Protocols | |
|---|---|
| IHC protocol for PDRG1 antibody 85851-4-RR | Download protocol |
| WB protocol for PDRG1 antibody 85851-4-RR | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |