Co-immunoprecipitation troubleshooting, tips and tricks
Co-immunoprecipitation (Co-IP) describes the isolation of a protein and its binding partners from a cell extract using a Nanobody or antibody that is bound to beads. The protein, that directly interacts with the Nanobody, or antibody beads is called “bait”. The binding partner that is indirectly precipitated is called “prey”.
Co-IP with GFP-Trap
In the following blog we will use ChromoTek’s GFP-Trap® as an example. GFP-Trap consists of an anti-GFP Nanobody conjugated to beads and is used to immunoprecipitate a GFP-tagged bait protein (light and dark green) and its interacting prey protein (red). Analysis of the Co-IP reaction is done by Western blot (WB) using an anti-GFP antibody such as ChromoTek GFP Monoclonal antibody (3H9) for detection of the GFP-bait protein and an anti-prey antibody for visualization of the prey.
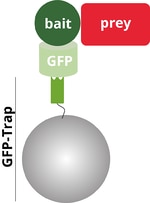
A complete Co-IP experiment also consists of positive and negative controls
In the recent blog “What is Co-immunoprecipitation (Co-IP)?” we described the workflow of Co-IP. We also explained in detail which controls are needed for meaningful results:
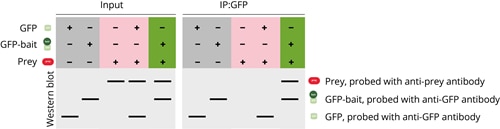
- Positive control: GFP and GFP-bait are analyzed in the absence of the prey protein. Precipitation of both GFP and GFP-bait confirms that the right conditions are chosen for the IP reaction.
- Negative control: The prey protein is analyzed in the absence of GFP-bait or with GFP only. The prey protein is not supposed to be precipitated when no bait protein is present.
- Co-IP experiment: The prey protein is analyzed in the presence of GFP-bait. The prey and GFP-bait protein are supposed to be precipitated together which confirms that they interact with each other.
A complete Co-IP experiment looks like this when analyzed on WB:
Troubleshooting cases
In the following section we discuss three frequently occurring Co-IP problems, i.e., “troubles”, using a setup comprising a GFP-tagged bait-protein, a prey protein, and GFP-Trap. Based on mandatory controls we show how the problems look like on WB, how to diagnose and finally how to resolve them. Although the presented Co-IP troubleshooting cases focus on GFP-Trap, the same principles also apply to tags other than GFP or for Co-IPs using conventional antibodies.
Case I: No pulldown of the GFP-bait protein
Case II: No pulldown of the prey protein
Case III: Unspecific pulldown of the prey protein
Case I: No pulldown of the GFP-bait protein
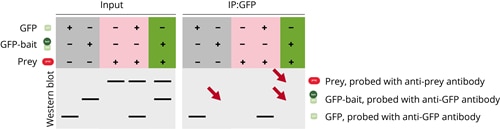
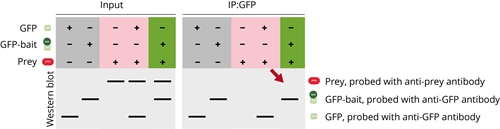
In the first case GFP-bait is present in the input fraction, but it isn’t precipitated and not found in the IP fraction. Consequently, the prey protein isn’t found in the IP fraction. The general IP setup seems to work, since the positive control where only GFP is precipitated is successful. It can be concluded that there is an issue with the precipitation of the GFP-bait protein.
Possible reasons are that the GFP-bait is not soluble after cell lysis or unfolded, which leads to immunoprecipitation of very small amounts of GFP-bait or to no precipitation of GFP-bait at all.
In this case it’s recommended to optimize the IP of the GFP-bait protein first. Different expression conditions for the GFP-bait, varying cell lysis and IP buffers as well as lysis and IP conditions should be tested. After having found the right conditions for the IP GFP-bait, Co-IP can be repeated in the presence of the GFP-bait and prey protein.
Case II: No pulldown of the prey protein
In the second case the prey protein is found in the input fraction, but its precipitation fails and can’t be found in the IP fraction. The precipitation of GFP and GFP-bait, however, works and both are detected in the IP fraction. This result indicates that there is an issue with the prey protein.
In such a case often the prey protein is unfolded, denatured and not soluble which prevent the prey protein interacting with the GFP-bait. It is also possible that the selected washing conditions are too harsh, and the prey is washed away.
Here it’s recommended to optimize the IP conditions for the prey protein by testing different expression conditions, modifying the lysis, IP and wash buffers, as well as lysis and IP conditions. It should also be considered whether there is really an interaction between the bait and prey or if e.g., the chosen tag of the bait protein could interfere with binding to the prey.
Case III: Unspecific pulldown of the prey protein
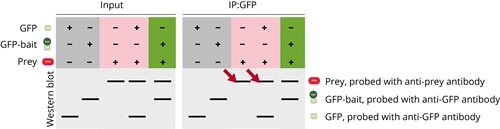
In the third case the prey protein is found in the IP fraction although no bait protein is present, for example when only GFP is used or when no GFP-bait is present. The prey protein is non-specifically precipitated by GFP-Trap.
Similar to case II the prey protein could be unfolded or denatured which leads to a non-specific interaction of the prey with the bead matrix, the Nanobody or plastic consumables.
During troubleshooting it’s necessary to find the optimal conditions for the prey protein. Often using a more stringent wash buffer and low binding plastic consumables can help to prevent non-specific binding of the prey. In addition, testing different expression conditions for prey, changing the lysis and IP buffers and lysis and IP conditions is recommended. The test should be conducted in the absence of the bait and once the optimal conditions are found, the Co-IP can be repeated with the GFP-bait protein.
Controls are essential for troubleshooting
Controls are not only essential for meaningful results, but they are also key for successful troubleshooting. Only when looking at the positive and negative controls together it’s possible to identify potential issues during troubleshooting and to interpret Co-IP results properly. Especially when conducting a Co-IP, controls are important to avoid false positive and false negative results and often optimization is required.
For IP and Co-IP troubleshooting, please also watch our webinar "immunoprecipitation (IP) troubleshooting online training".
For more details on Co-IP, please see our blog "How to conduct a Co-immunoprecipitation".
