Detecting RNA Methylation by Dot Blotting
A step-by-step guide by Proteintech scientists!
Jump to Protocol
What is an RNA dot blot?
An RNA dot blot is a technique that allows for the detection and semi-quantification of RNA molecules in a sample. The technique can also be used to detect RNA modifications, including RNA methylation.
How does an RNA dot blot work?
The protocol for RNA dot blotting is similar to northern blotting except RNA samples are not separated by electrophoresis. Instead, extracted RNA is directly spotted onto a nitrocellulose (NC) or nylon membrane followed by hybridization with a radioactive probe to detect the target RNA sequences. Alternatively, traditional blotting techniques using primary and secondary antibody incubation can also be used to detect post-transcriptional modifications to RNA sequences, such as RNA methylation.
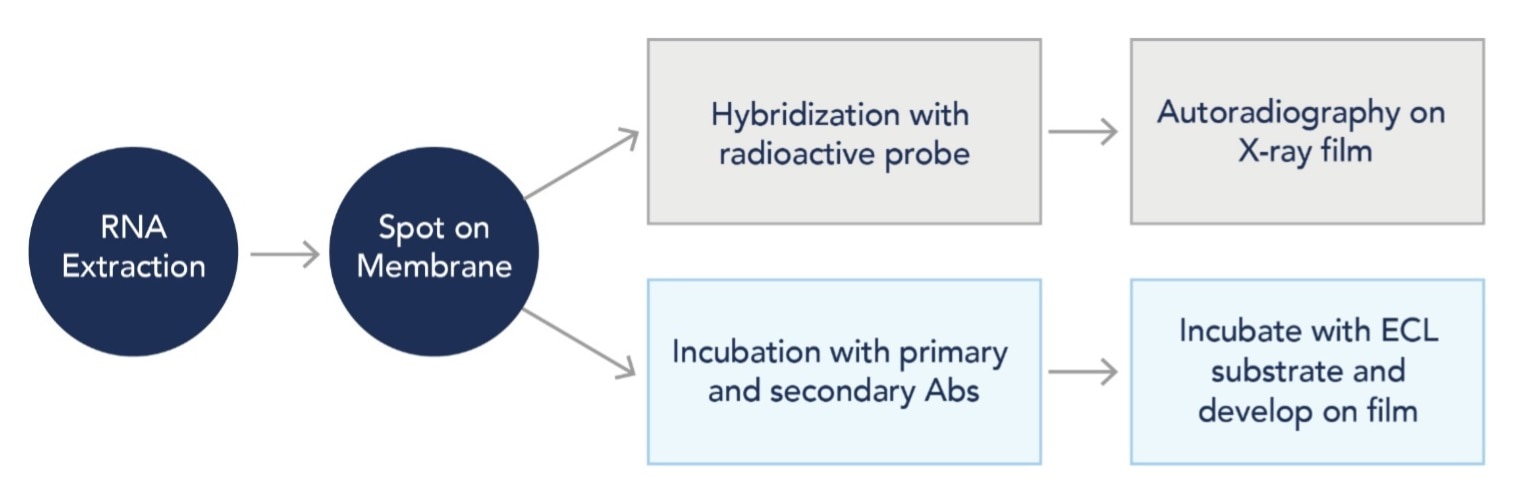
Figure 1: Simplified examples of RNA dot blot workflows
Why is dot blotting a good technique to observe RNA methylation?
RNA methylation is a post-transcriptional modification that adds a methyl group (-CH3) to specific RNA nucleotides. These modifications play critical roles in many biological processes, including RNA metabolism and gene expression, development and differentiation, and disease. N6-methyladenosine (m6A) is the most common RNA modification in eukaryotes. Other types of RNA methylation include N1-methyladenosine (m1A), 5-methylcytosine (m5C), and 7-methylguanosine (m7G).
By using antibodies designed to recognize methylated RNA molecules, dot blotting can be a powerful tool for analyzing RNA methylation for several reasons:
Sensitivity: Dot blotting is highly sensitive and can detect RNA methylation at very low levels. Since RNA methylation is often low in RNA extracts, highly sensitive techniques are required to detect it.
Simplicity: Dot blotting is a relatively simple technique that requires minimal sample preparation and equipment. It does not require RNA sequencing or other advanced techniques, making it a cost-effective and efficient option for detecting RNA methylation.
Specificity: Dot blotting can be highly specific for detecting RNA methylation if a specific antibody or probe is used. This can be important for distinguishing RNA methylation from other post-transcriptional modifications or products from RNA degradation.
High-throughput: Dot blotting can be easily adapted for high-throughput screening of a large number of samples. This makes it a useful technique for large-scale studies of RNA methylation in various biological contexts.
It is important to note that dot blotting is a semi-quantitative technique, and quantitative measurements of RNA modifications may require other techniques such as RNA sequencing or mass spectrometry.
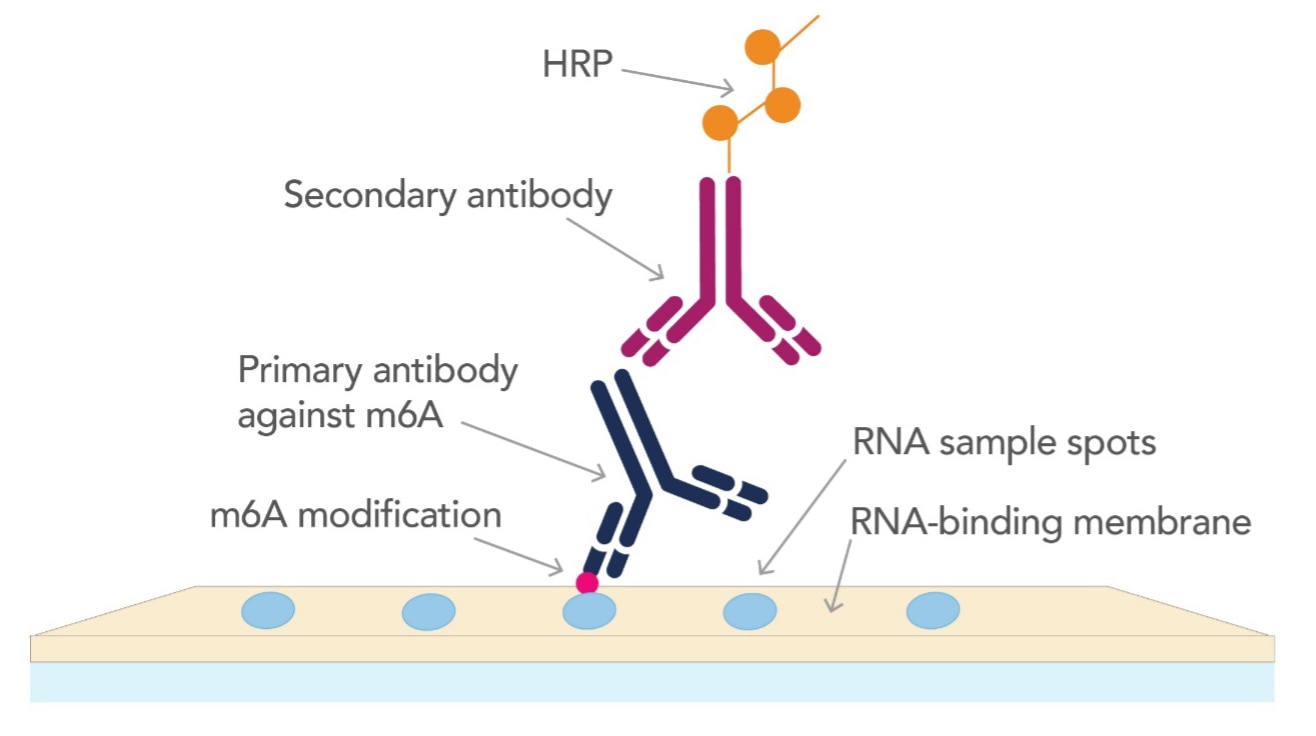
Figure 2: Graphical representation of identifying m6A modification by dot blot
Proteintech offers RNA methylation antibodies validated for dot blot!
m6A Monoclonal Antibody (68055-1-lg)
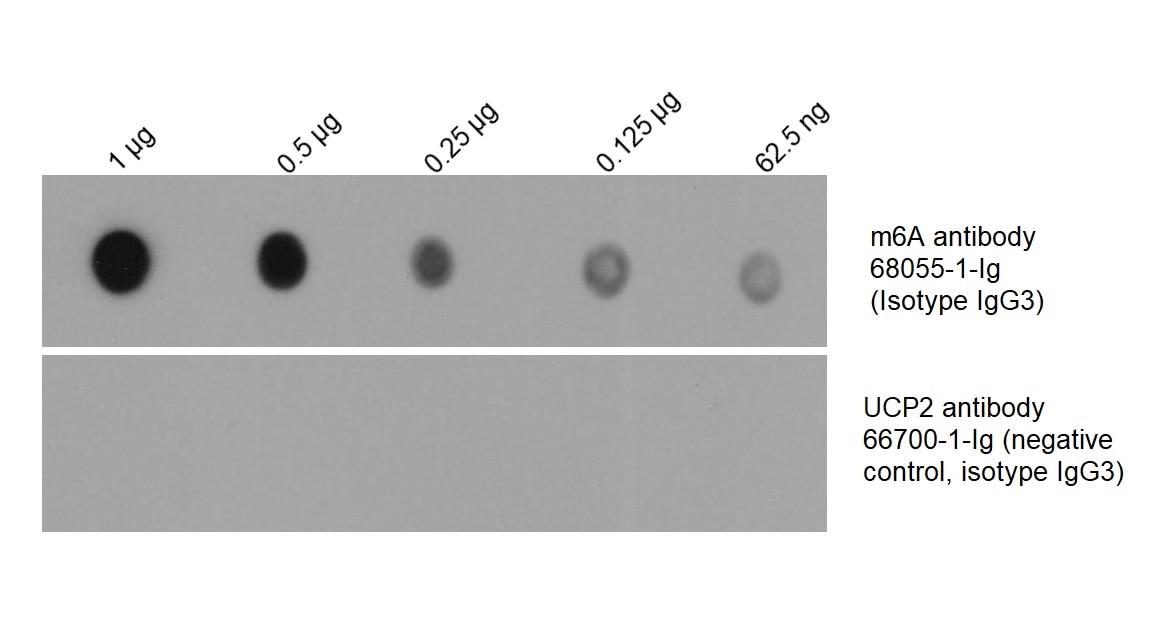
Figure 3: Total RNA was isolated from HEK-293 cells and dotted on an NC membrane at different amounts. The membrane was blotted with m6A antibody (68055-1-Ig, 1:2000) followed by incubation with HRP-goat anti-mouse secondary antibody. A parallel dot blot was performed using an antibody with the same isotype (UCP2 antibody 66700-1-Ig) at the same dose.
m5C Monoclonal Antibody (68301-1-lg)
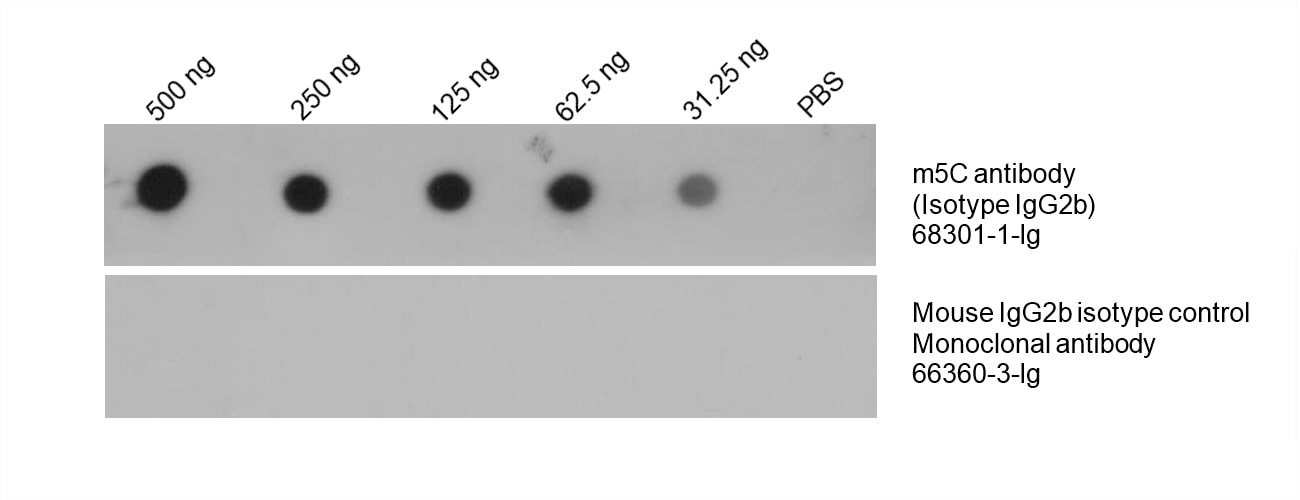 Figure 4: Total DNA was isolated from HeLa cell line and dotted on an NC membrane at different amounts. The membrane was blotted with m5C antibody (68301-1-Ig, 1:5000) followed by incubation of HRP-goat anti-mouse secondary antibody. A parallel dot blot was performed using Mouse IgG2b isotype control Monoclonal antibody 66360-3-Ig at the same dose.
Figure 4: Total DNA was isolated from HeLa cell line and dotted on an NC membrane at different amounts. The membrane was blotted with m5C antibody (68301-1-Ig, 1:5000) followed by incubation of HRP-goat anti-mouse secondary antibody. A parallel dot blot was performed using Mouse IgG2b isotype control Monoclonal antibody 66360-3-Ig at the same dose.
m7G Monoclonal Antibody (68302-1-lg)
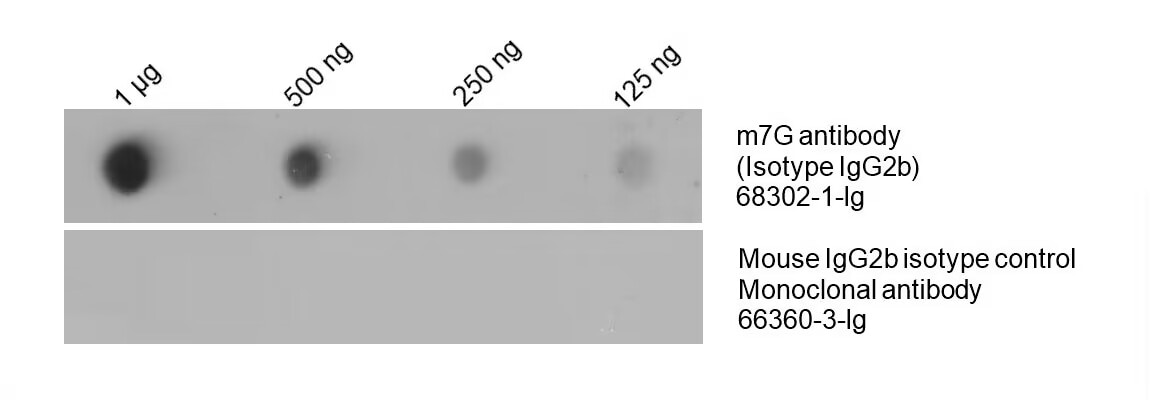
Figure 5: Total RNA was isolated from HeLa cell line and dotted on an NC membrane at different amounts. The membrane was blotted with m7G antibody (68302-1-Ig, 1:5000) followed by incubation of HRP-goat anti-mouse secondary antibody. A parallel dot blot was performed using Mouse IgG2b isotype control Monoclonal antibody 66360-3-Ig at the same dose.
Protocol for RNA dot blot using antibodies for RNA methylation
- Extract RNA from samples with extraction reagent (or kit) and formulate a proper final concentration between 0.05-2 μg/μL. Dilute with PBS if the concentration is too high. Avoid any contamination with RNase.
- Directly dot the extracted RNA onto the nitrocellulose (NC) membrane. Mark the dot position and front side of the membrane. The appropriate amount of RNA for each dot is between 0.2–2 μg. Serial dilutions are recommended to determine the optimal amount. Adjust each dot to the same volume by dilution with PBS if needed.
- Dry the membrane for 5–15 minutes at room temperature, and block with 1% BSA (in PBST) solution for one hour. If methylene blue staining is applied as a loading control, the dried membrane can be directly stained without blocking. Blocking will interfere with methylene blue staining.
- Briefly wash with PBS, PBST, or TBST once. Dilute the primary antibody with 1% BSA (in PBST) solution. The following dilutions are recommended for Proteintech’s m6A, m5C, and m7G antibodies:
m6A Monoclonal Antibody (68055-1-Ig): 1:2000
m5C Monoclonal Antibody (68301-1-Ig): 1:5000
m7G Monoclonal Antibody (68302-1-Ig): 1:5000
Add the diluted primary antibody solution to the membrane and incubate at room temperature for 1.5 hours. Ensure that the primary antibody solution completely covers the membrane. Avoid using milk for dilution.
- Wash with PBST 4 times for 5 minutes each time. Discard the washing buffer, add the secondary antibody diluted in 1% BSA, and incubate at room temperature for 1.5 hours or at 37℃ for 1 hour. Ensure that the secondary antibody solution completely covers the membrane. Avoid using milk for dilution.
- Wash in PBST 4 times for 5 minutes each time. Completely discard the washing buffer and add ECL substrate for the luminescent development.
Related Content
New avenue for treatment of ALS and frontotemporal dementia
7 Tech Tips for Successful ChIP Experiments
ChIP with RFP-tagged or mCherry -tagged proteins
Support
Newsletter Signup
Stay up-to-date with our latest news and events. New to Proteintech? Get 10% off your first order when you sign up.