Proteases for Western Blotting: Choosing the right tools for the job
How proteases can help you to better understand your Western blot results
Proteases are often labeled as “bad guys” that complicate our Western blot analyses. They cut up target proteins into unrecognizable pieces that do not resemble the expected results. However, utilising specific proteases in your experimental design can help you to further understand the molecular weight of your observed bands.
There are two types of enzymes that can be helpful: phosphatase and glycosidase.
Phosphatase enzymes
Let’s start with phosphatase. In the study of signaling pathways, protein phosphorylation and dephosphorylation are ubiquitous. Protein phosphorylation is catalyzed by a protein kinase, and dephosphorylation is catalyzed by phosphatase.
In WB analysis of phosphorylated proteins, there will often be two close bands near the predicted molecular weight. To test whether the upper band is the phosphorylated form, phosphatase can be used. The disappearance or weakening of the upper band – but not the lower band – indicates that the upper band is a phosphorylated form of this protein.
Proteintech always works with phosphatases to verify the specificity of antibodies targeting phosphorylated proteins. An example is our RIPK1 Phospho-S161 monoclonal antibody (66854-1-IG). In the validation data below (Figure 1), the signal in several cell lysates disappeared after phosphatase treatment.
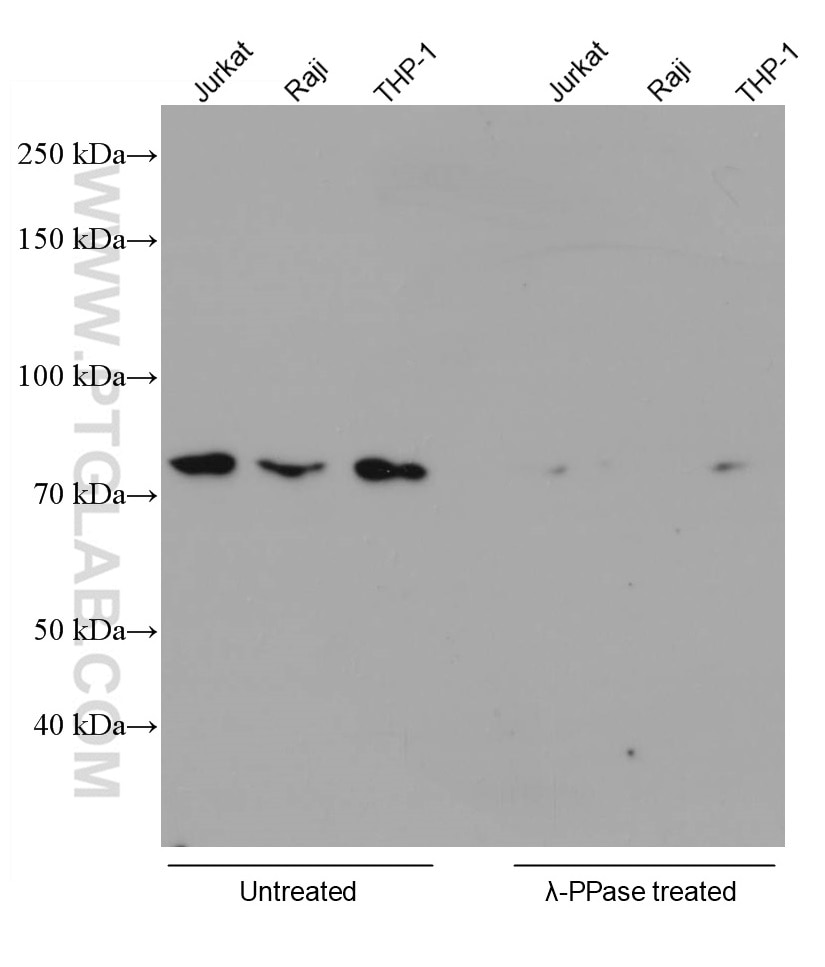 |
Figure 1. Various lysates were subjected to SDS PAGE followed by WB with 66854-1-Ig (RIPK1 phospho-S161 antibody) at a dilution of 1:5000 incubated at room temperature for 1.5 hours. The cell lysates were untreated (left) or treated with Lambda Protein Phosphatase (lamda-PPase, 500U, right) at 37oC for 1 hour before the blocking step. |
A word of caution here: many commercially available phosphatases are inactivated by lysis buffer. We recommend checking with the manufacturer before using them in your experiments.
Glycosidase enzymes
In Western blot, we often encounter proteins (especially membrane proteins) where the observed molecular weight is much larger than the calculated value. One possible cause of this discrepancy is glycosylation. Applying a glycosidase enzyme treatment to your cells can help you determine whether the observed higher molecular weight is due to glycosylation: if the band appears at the expected weight following glycosidase treatment, the target is likely a glycoprotein.
There are three commonly used glycosidase enzymes:
- PNGase F – Cuts off almost all types of asparagine-linked carbohydrates.
- EndoH – Cuts high mannose and some hybrid types of N-linked carbohydrates.
- O-Glycosidase – Cuts off O-linked carbohydrates from serine and threonine residues.
Take our CD2 monoclonal antibody (60005-1-IG), for example. CD2 is composed of 351 amino acids and the calculated size is 39kDa. However, the target migrates to ~50kDa in its glycosylated form. To confirm that the deviation is due to glycosylation, we used the glycosidase PNGase F, which cleaves asparagine-linked glycosyl groups from proteins. Below are the results using Jurkat cells (Figure 2). After PNGase F treatment, the target protein migrates to near the calculated value.
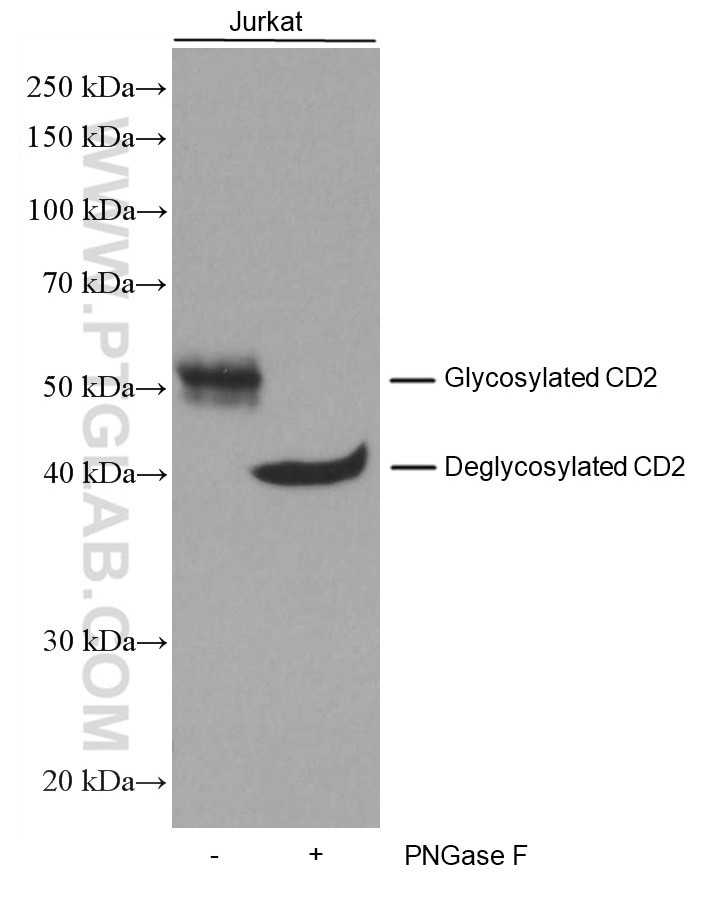 |
Figure 2. Untreated and PNGase F-treated Jurkat cell lysates were subjected to SDS PAGE followed by WB with 60005-1-Ig (CD2 antibody) at a dilution of 1:3000 incubated at room temperature for 1.5 hours. |
Table 1. Common troubleshooting issues in glycosylase experiments
Co
| Issue | Possible causes | Solution |
|
No change after enzyme digestion |
1. The enzyme chosen is not compatible with the lysis buffer. 2. Insufficient enzyme amount or incubation time. 3. The detection resolution is not enough. 4. The target protein is unmodified. |
1. Ensure compatibility with enzyme manufacturer. 2. Conduct trials with varying amounts of these parameters. 3. Use an appropriate gel concentration to resolve close bands. 4. Combine with other experiments to confirm the presence or absence of modification. |
|
Putative glycosylated species weakens only slightly after digestion |
1. Insufficient enzyme amount or incubation time. 2. The target signal is produced by two proteins or two forms of the same protein. |
1. Increase the amount of enzyme used or extend the incubation time. 2. Combine other experiments to confirm the presence of other modifications or non-related protein interference. |
|
The size of the enzyme after digestion does not match the calculated value |
1. Insufficient enzyme amount or incubation time. 2. There are other modifications to the protein. |
1. Increase the amount of enzyme used or extend the enzyme cutting time. 2. Verify other modifications in conjunction with other experiments. |
Related Products
Western blot products overview
Tag & loading control antibodies
SignalBright chemiluminescent substrate
Related Content
Detecting low abundance proteins via Western blot
In search of low molecular weight proteins
Lysate preparation: why RIPA buffer is best
Lysate Preparation: How Do I Optimize My Extraction?
Loading control antibodies for Western blot
Comparing BCA, Bradford, and UV protein quantification methods for scientists
Why does the observed protein molecular weight (MW) differ from the calculated one?
Learn how to save precious hours on your IP, IF, and Western blotting experiments

Support
Newsletter Signup
Stay up-to-date with our latest news and events. New to Proteintech? Get 10% off your first order when you sign up.