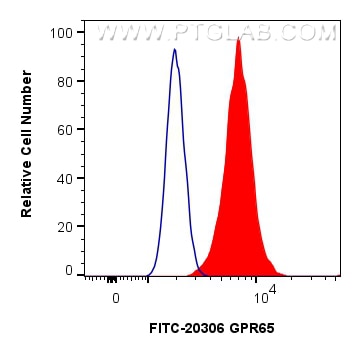
Tested Applications
| Positive FC (Intra) detected in | RAW 264.7 cells |
Recommended dilution
| Application | Dilution |
|---|---|
| Flow Cytometry (FC) (INTRA) | FC (INTRA) : 0.8 ug per 10^6 cells in a 100 µl suspension |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
FITC-20306 targets GPR65 in FC (Intra) applications and shows reactivity with human samples.
| Tested Reactivity | human |
| Host / Isotype | Rabbit / IgG |
| Class | Polyclonal |
| Type | Antibody |
| Immunogen |
CatNo: Ag14139 Product name: Recombinant human GPR65 protein Source: e coli.-derived, PGEX-4T Tag: GST Domain: 238-337 aa of BC035633 Sequence: CFTPFHVMLLIRCILEHAVNFEDHSNSGKRTYTMYRITVALTSLNCVADPILYCFVTETGRYDMWNILKFCTGRCNTSQRQRKRILSVSTKDTMELEVLE Predict reactive species |
| Full Name | G protein-coupled receptor 65 |
| Calculated Molecular Weight | 337 aa, 39 kDa |
| GenBank Accession Number | BC035633 |
| Gene Symbol | GPR65 |
| Gene ID (NCBI) | 8477 |
| Conjugate | FITC Fluorescent Dye |
| Excitation/Emission Maxima Wavelengths | 498 nm / 526 nm |
| Excitation Laser | Blue laser (488 nm) |
| Form | Liquid |
| Purification Method | Antigen affinity purification |
| UNIPROT ID | Q8IYL9 |
| Storage Buffer | PBS with 50% glycerol, 0.05% Proclin300, 0.5% BSA, pH 7.3. |
| Storage Conditions | Store at -20°C. Avoid exposure to light. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
Background Information
GPR65 (G protein-coupled receptor 65) was defined originally as T-cell death-associated gene 8 (TDAG8), functions as a proton-sensing receptor, and is also sensitive to psychosine (PMID: 35343079). GPR65 promotes adaptation to an acidic environment to enhance cell survival and proliferation, thereby promoting tumor development (PMID: 36852075).
Protocols
| Product Specific Protocols | |
|---|---|
| FC protocol for FITC GPR65 antibody FITC-20306 | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |