Tested Applications
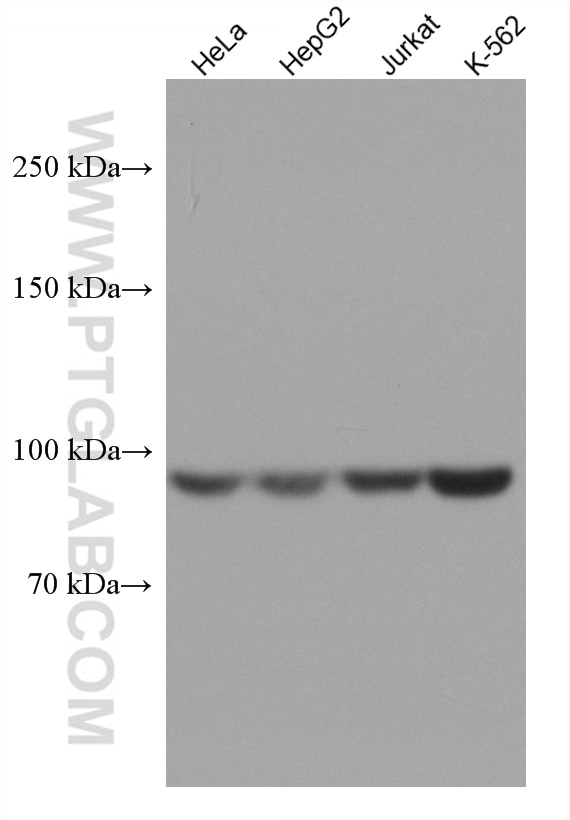
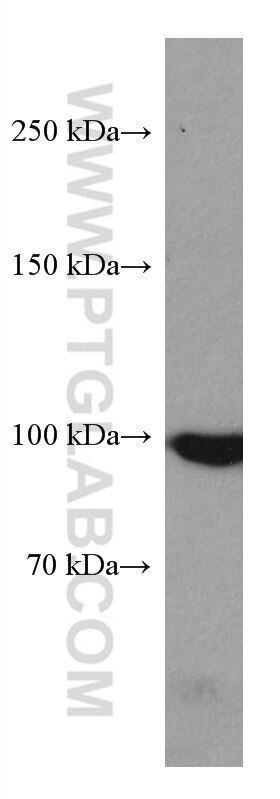
| Positive WB detected in | HeLa cells, A431 cells, HepG2 cells, Jurkat cells, K-562 cells |
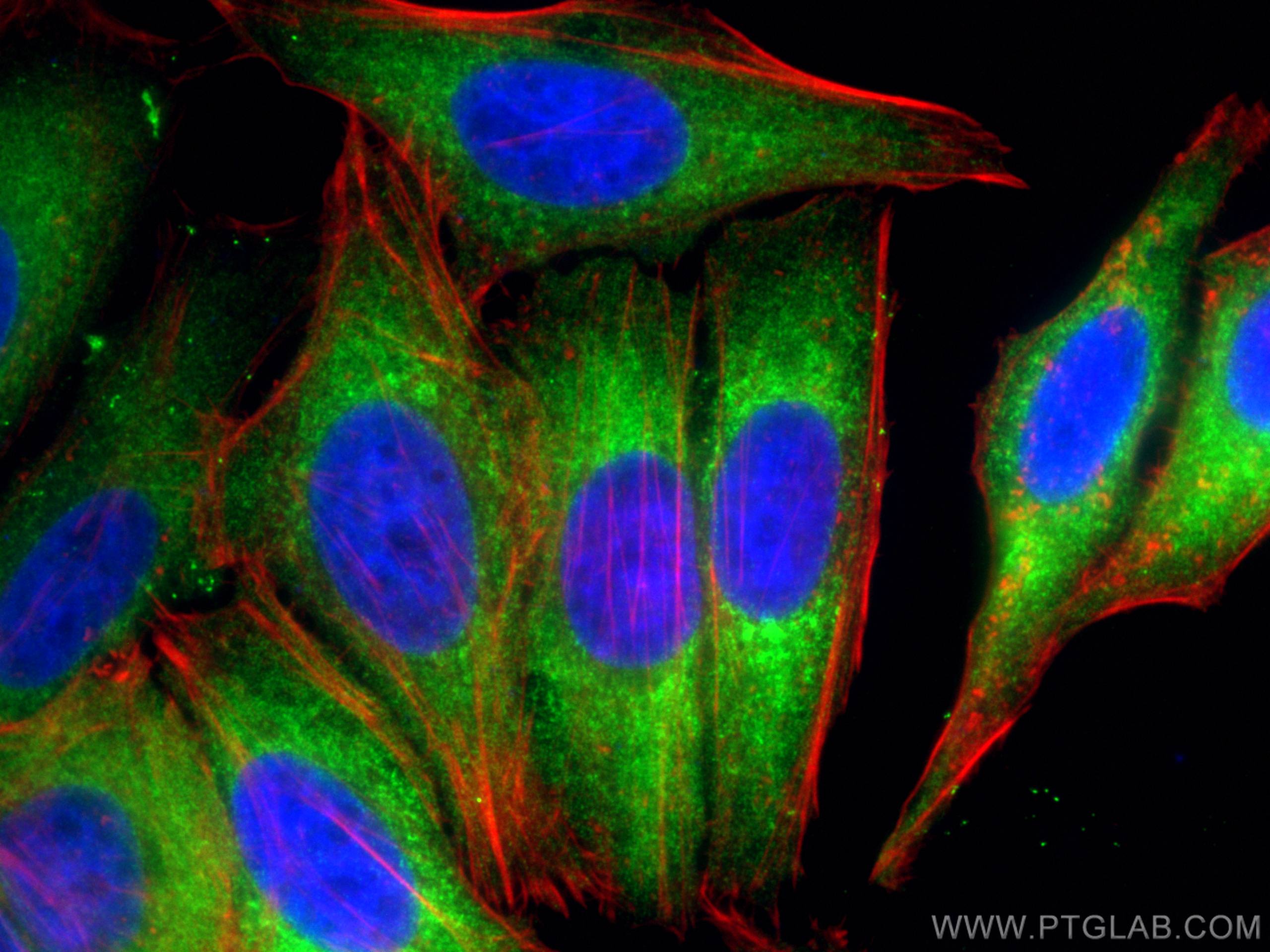
| Positive IF/ICC detected in | HepG2 cells |
Recommended dilution
| Application | Dilution |
|---|---|
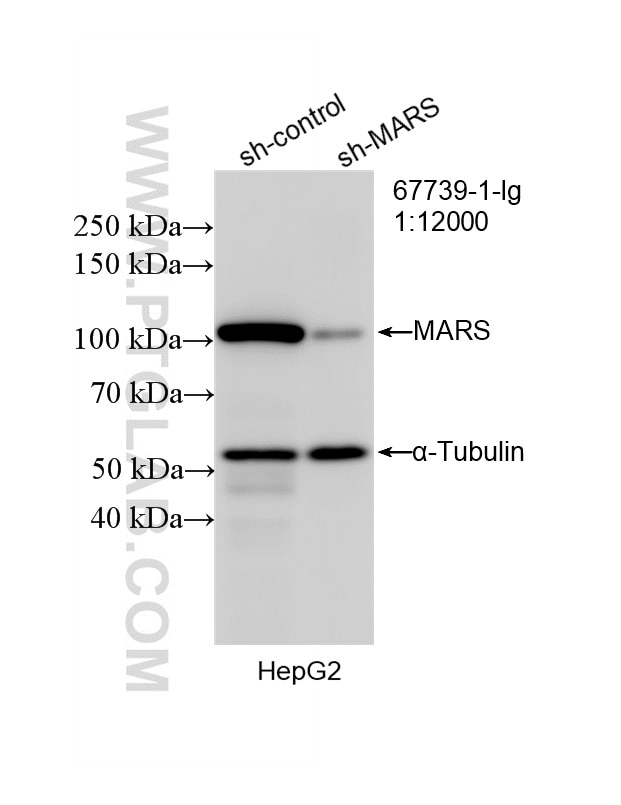
| Western Blot (WB) | WB : 1:5000-1:50000 |
| Immunofluorescence (IF)/ICC | IF/ICC : 1:200-1:800 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
67739-1-Ig targets MARS in WB, IF/ICC, ELISA applications and shows reactivity with human samples.
| Tested Reactivity | human |
| Host / Isotype | Mouse / IgG2a |
| Class | Monoclonal |
| Type | Antibody |
| Immunogen |
CatNo: Ag6961 Product name: Recombinant human MARS protein Source: e coli.-derived, PET28a Tag: 6*His Domain: 552-900 aa of BC002384 Sequence: MAKDNVPFHSLVFPCSALGAEDNYTLVSHLIATEYLNYEDGKFSKSRGVGVFGDMAQDTGIPADIWRFYLLYIRPEGQDSAFSWTDLLLKNNSELLNNLGNFINRAGMFVSKFFGGYVPEMVLTPDDQRLLAHVTLELQHYHQLLEKVRIRDALRSILTISRHGNQYIQVNEPWKRIKGSEADRQRAGTVTGLAVNIAALLSVMLQPYMPTVSATIQAQLQLPPPACSILLTNFLCTLPAGHQIGTVSPLFQKLENDQIESLRQRFGGGQAKTSPKPAVVETVTTAKPQQIQALMDEVTKQGNIVRELKAQKADKNEVAAEVAKLLDLKKQLAVAEGKPPEAPKGKKKK Predict reactive species |
| Full Name | methionyl-tRNA synthetase |
| Calculated Molecular Weight | 101 kDa |
| Observed Molecular Weight | 97-101 kDa |
| GenBank Accession Number | BC002384 |
| Gene Symbol | MARS |
| Gene ID (NCBI) | 4141 |
| RRID | AB_2918508 |
| Conjugate | Unconjugated |
| Form | Liquid |
| Purification Method | Protein A purification |
| UNIPROT ID | P56192 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol, pH 7.3. |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. 20ul sizes contain 0.1% BSA. |
Protocols
| Product Specific Protocols | |
|---|---|
| IF protocol for MARS antibody 67739-1-Ig | Download protocol |
| WB protocol for MARS antibody 67739-1-Ig | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |