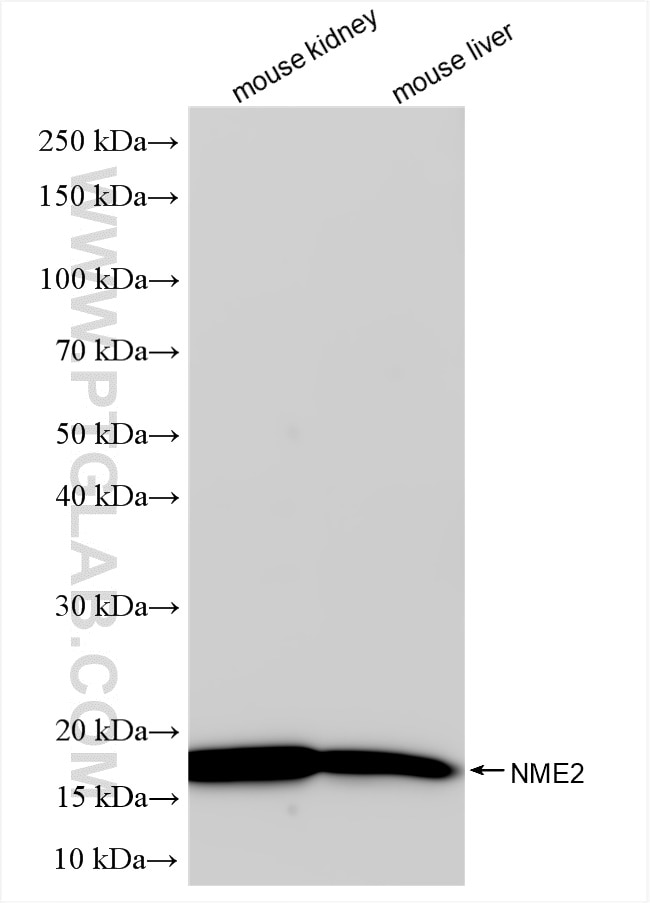
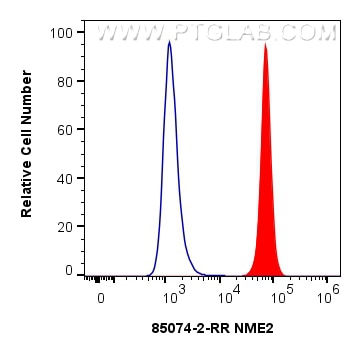
Tested Applications
| Positive WB detected in | mouse kidney tissue, mouse liver tissue |
| Positive FC (Intra) detected in | HEK-293T cells |
Recommended dilution
| Application | Dilution |
|---|---|
| Western Blot (WB) | WB : 1:5000-1:50000 |
| Flow Cytometry (FC) (INTRA) | FC (INTRA) : 0.25 ug per 10^6 cells in a 100 µl suspension |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
85074-2-RR targets NME1/2 in WB, FC (Intra), ELISA applications and shows reactivity with human, mouse samples.
| Tested Reactivity | human, mouse |
| Host / Isotype | Rabbit / IgG |
| Class | Recombinant |
| Type | Antibody |
| Immunogen |
CatNo: Ag13913 Product name: Recombinant human NME2 protein Source: e coli.-derived, PGEX-4T Tag: GST Domain: 1-152 aa of BC002476 Sequence: MANLERTFIAIKPDGVQRGLVGEIIKRFEQKGFRLVAMKFLRASEEHLKQHYIDLKDRPFFPGLVKYMNSGPVVAMVWEGLNVVKTGRVMLGETNPADSKPGTIRGDFCIQVGRNIIHGSDSVKSAEKEISLWFKPEELVDYKSCAHDWVYE Predict reactive species |
| Full Name | non-metastatic cells 2, protein (NM23B) expressed in |
| Calculated Molecular Weight | 152 aa, 17 kDa |
| Observed Molecular Weight | 17 kDa |
| GenBank Accession Number | BC002476 |
| Gene Symbol | NME2 |
| Gene ID (NCBI) | 4831 |
| Conjugate | Unconjugated |
| Form | Liquid |
| Purification Method | Protein A purification |
| UNIPROT ID | P22392 |
| Storage Buffer | PBS with 0.02% sodium azide and 50% glycerol, pH 7.3. |
| Storage Conditions | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. 20ul sizes contain 0.1% BSA. |
Background Information
NME2(non-metastatic cells 2, protein) is also named as NDKB(Nucleoside diphosphate kinase B), NM23B and belongs to the NDK family. It is involved in the phosphorylation of nucleoside diphosphates and interacts with membrane receptor and constituting a novel molecular regulatory mechanism of GPCR endocytosis. This protein has 2 isoforms produced by alternative splicing. Refer to epitope and protein detection, the antibody 85074-2-RR detects both NME1& NME2.
Protocols
| Product Specific Protocols | |
|---|---|
| FC protocol for NME1/2 antibody 85074-2-RR | Download protocol |
| WB protocol for NME1/2 antibody 85074-2-RR | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |