5 Tips for better immunoprecipitation (IP)
Pull-down of proteins can be difficult, particularly when they are expressed at low levels. Here we present 5 tips, which will considerably improve your IP results.
- Protein concentration matters
- Binding affinity matters
- Time matters
- Specificity matters
- Volume matters
- Free high affinity Nano-Trap sample
The higher the protein concentration, the higher the IP yield. Try to use a concentration as high as possible.It makes a significant difference if the concentration of your protein during IP is 1 nM (low protein expression level), about 50 nM (intracellular endogenous protein expression level, will be diluted by buffer for IP) or 1000 nM (high protein concentration).
Same protein input, different KD affinity matrices resulting in different yields!
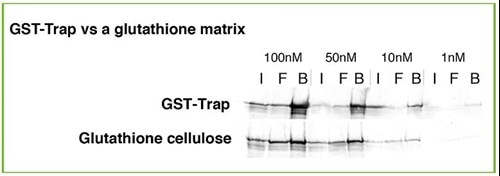
The higher the binding affinity, the higher the protein yield.
A specific antibody, which works well using high abundant proteins, may perform poor when low abundant proteins shall be precipitated. Make sure that your antibody has a low dissociation constant KD respectively high affinity. Otherwise your antibody binds just a very small fraction of your protein. At a protein concentration of 50 nM the IP with a low affinity media (KD of 150 nM) will bind only 25% at best, whereas a high affinity matrix (KD of 1nM) will bind 98% of the protein. This is a critical difference!
|
Affinity matrix |
KD |
Fraction of bound protein in % & protein yield in pmol at given protein concentration |
||
|
1 nM |
10 nM |
50 nM |
||
|
Protein yield of affinity media |
150 nM |
<1 % |
6 % |
25 % |
|
<0.005 pmol |
0.3 pmol |
6.25 pmol |
||
|
Immunoprecipitation using high affinity matrix, e.g. Nano-Trap |
1 nM |
50 % |
91 % |
98 % |
|
0.25 pmol |
4.55 pmol |
24.5 pmol |
||
Pull down or immunoprecipitation: The fraction and amount of protein captured by an affinity matrix or antibody is dependent on protein concentration and on the dissociation constant KD of the matrix-protein interaction. Protein amounts stated for a total volume of 500 µl (sample, buffer, and bead slurry) used in IP.
For instance, the ChromoTek GFP-Trap has a very low KD of 1 pM (high affinity) and enables very efficient immunoprecipitations of very low protein concentrations of GFP-fusion proteins.
The shorter the wash time, the better the pull-down yield. The optimal wash time has to be determined experimentally (also consider background). However, the use of affinity media with low off rates helps to avoid leaching of your bound protein.
The better the specificity of the affinity matrix, the lower the background. Use only specific, well-characterized, and validated affinity matrices to obtain reliable, reproducible results.
The smaller the volume, the more effective your IP works. Using a small volume keeps your protein concentration high and therefore increases the binding affinity. Concentration is a function of volume. Try to use a volume as small as possible. That keeps your protein concentration high and therefore increases the binding affinity.
In summary, we recommend keeping protein concentration, binding affinity, processing time, IP resin’s specificity, and buffer volume in mind when planning your next immunoprecipitation to obtain the best immunoprecipitation results.
You can test Nano-Traps with high binding affinities yourself. Just request a free sample here:
For a detailed explanation and description, download our white paper Immunoprecipitation of proteins at very low expression levels using GST-Trap and MBP-Trap.
Here we demonstrate the superior performance of the ChromoTek Nano-Traps affinity media using GST-Trap and MBP-Trap and present a theoretical framework how their low dissociation constants translate into effective immunoprecipitation of proteins expressed at low levels or from dilute solutions.
