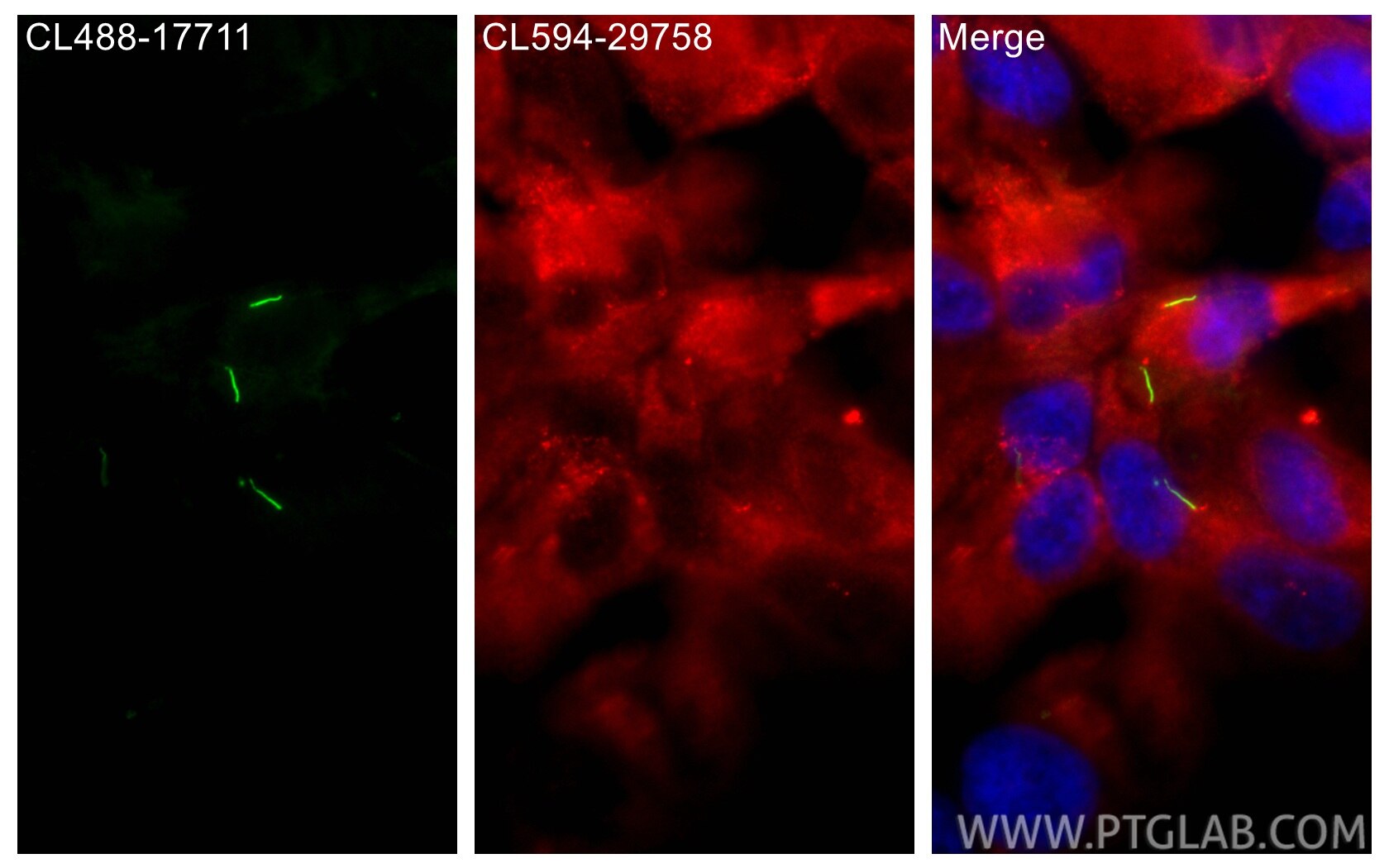
Tested Applications
| Positive IF/ICC detected in | hTERT-RPE1 cells |
Recommended dilution
| Application | Dilution |
|---|---|
| Immunofluorescence (IF)/ICC | IF/ICC : 1:50-1:500 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
CL594-29758 targets DYNC2H1 in IF/ICC applications and shows reactivity with human, mouse samples.
| Tested Reactivity | human, mouse |
| Host / Isotype | Rabbit / IgG |
| Class | Polyclonal |
| Type | Antibody |
| Immunogen |
CatNo: Ag30707 Product name: Recombinant human DYNC2H1 protein Source: e coli.-derived, PGEX-4T Tag: GST Domain: 42-196 aa of NM_001377 Sequence: FLDDGNQMLLRVQRSDAGISFSNTIEFGDTKDKVLVFFKLRPEVITDENLHDNILVSSMLESPISSLYQAVRQVFAPMLLKDQEWSRNFDPKLQNLLSELEAGLGIVLRRSDTNLTKLKFKEDDTRGILTPSDEFQFWIEQAHRGNKQISKERAN Predict reactive species |
| Full Name | dynein, cytoplasmic 2, heavy chain 1 |
| Calculated Molecular Weight | 493 kDa |
| Observed Molecular Weight | 493 kDa |
| GenBank Accession Number | NM_001377 |
| Gene Symbol | DYNC2H1 |
| Gene ID (NCBI) | 79659 |
| Conjugate | CoraLite®594 Fluorescent Dye |
| Excitation/Emission Maxima Wavelengths | 588 nm / 604 nm |
| Form | Liquid |
| Purification Method | Antigen affinity purification |
| UNIPROT ID | Q8NCM8 |
| Storage Buffer | PBS with 50% glycerol, 0.05% Proclin300, 0.5% BSA, pH 7.3. |
| Storage Conditions | Store at -20°C. Avoid exposure to light. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
Background Information
DYNC2H1 (Cytoplasmic dynein 2 heavy chain 1) is involved in ciliary intraflagellar transport (IFT). DYNC2H1 drives retrograde transport of the IFT-A protein complex that regulates tip-to-base transport in cilia, involved in the generation and maintenance of cilia. DYNC2H1 variants or retina-predominant variants cause nonsyndromic retinal degeneration (PMID:32753734). DYNC2H1 mutations also cause asphyxiating thoracic dystrophy and short rib-polydactyly syndrome (PMID: 19442771). DYNC2H1 was localized to the basal bodies by immunofluorescence staining (PMID: 21552265, PMID: 26077881, PMID: 30320547).
Protocols
| Product Specific Protocols | |
|---|---|
| IF protocol for CL594 DYNC2H1 antibody CL594-29758 | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |