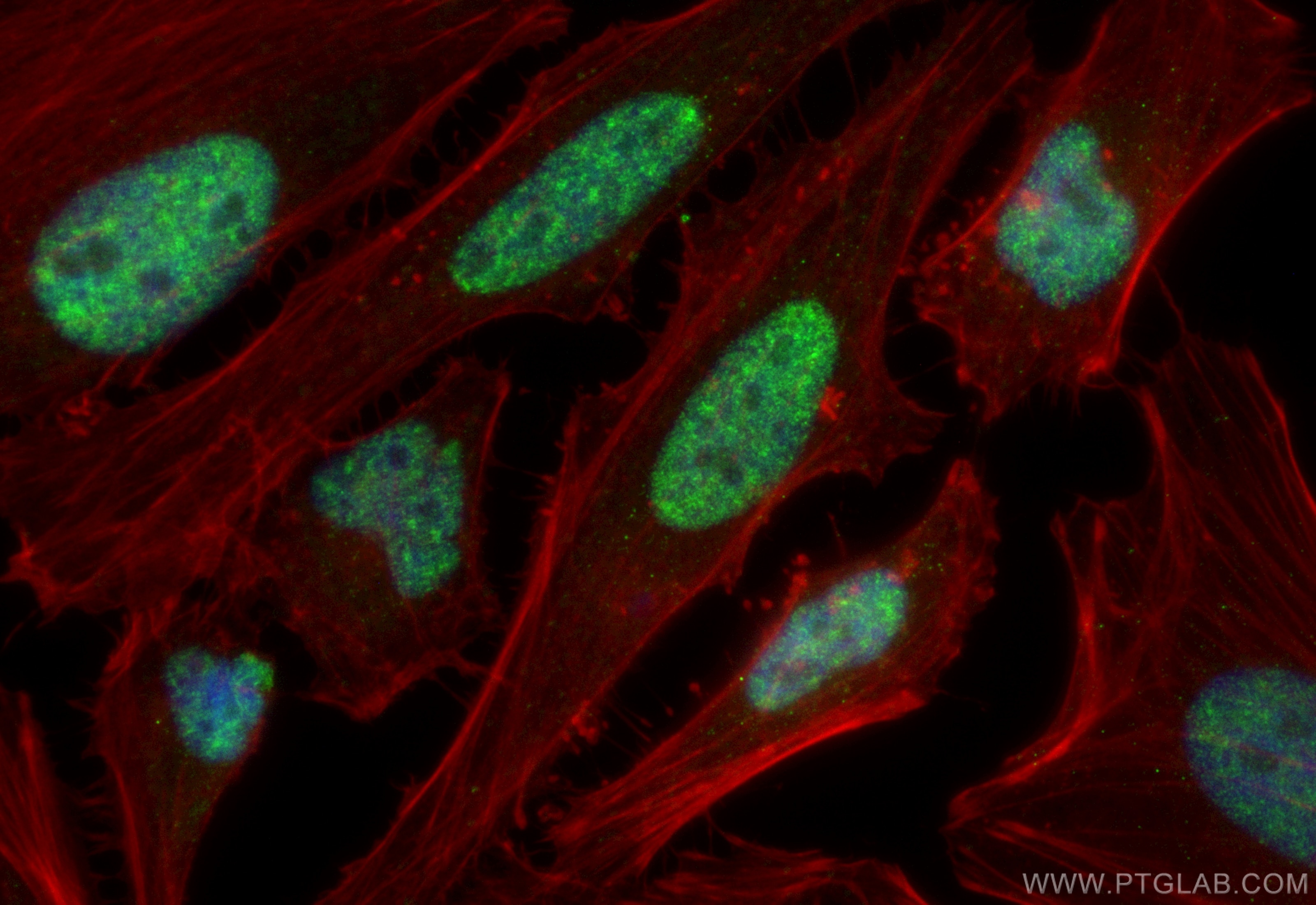
Tested Applications
| Positive IF/ICC detected in | HeLa cells |
Recommended dilution
| Application | Dilution |
|---|---|
| Immunofluorescence (IF)/ICC | IF/ICC : 1:50-1:500 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
Product Information
CL488-84491-6 targets PQBP1 in applications and shows reactivity with human samples.
| Tested Reactivity | human |
| Host / Isotype | Rabbit / IgG |
| Class | Recombinant |
| Type | Antibody |
| Immunogen |
CatNo: Ag9329 Product name: Recombinant human PQBP1 protein Source: e coli.-derived, PGEX-4T Tag: GST Domain: 1-265 aa of BC012358 Sequence: MPLPVALQTRLAKRGILKHLEPEPEEEIIAEDYDDDPVDYEATRLEGLPPSWYKVFDPSCGLPYYWNADTDLVSWLSPHDPNSVVTKSAKKLRSSNADAEEKLDRSHDKSDRGHDKSDRSHEKLDRGHDKSDRGHDKSDRDRERGYDKVDRERERDRERDRDRGYDKADREEGKERRHHRREELAPYPKSKKAVSRKDEELDPMDPSSYSDAPRGTWSTGLPKRNEAKTGADTTAAGPLFQQRPYPSPGAVLRANAEASRTKQQD Predict reactive species |
| Full Name | polyglutamine binding protein 1 |
| Calculated Molecular Weight | 265 aa, 30 kDa |
| Observed Molecular Weight | 30-32 kDa |
| GenBank Accession Number | BC012358 |
| Gene Symbol | PQBP1 |
| Gene ID (NCBI) | 10084 |
| Conjugate | CoraLite® Plus 488 Fluorescent Dye |
| Excitation/Emission Maxima Wavelengths | 493 nm / 522 nm |
| Form | Liquid |
| Purification Method | Protein A purification |
| UNIPROT ID | O60828 |
| Storage Buffer | PBS with 50% glycerol, 0.05% Proclin300, 0.5% BSA, pH 7.3. |
| Storage Conditions | Store at -20°C. Avoid exposure to light. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
Background Information
PQBP1 (Polyglutamine-binding protein 1) is also named as NPW38 and JM26. PQBP1 senses extrinsic tau 3R/4R proteins by direct interaction and triggers an innate immune response by activating a cyclic GMP-AMP synthase (cGAS)-Stimulator of interferon genes (STING) pathway (PMID: 34782623). PQBP1 is abundantly expressed in the central nervous system during development. PQBP1 is a component of the precatalytic spliceosome (B complex) that is involved in splicing. PQBP1 was predominantly localized in the cell nucleus. PQBP1 is overexpressed in various cancer types including breast cancer, colon cancer, and glioblastoma (PMID: 38342602).
Protocols
| Product Specific Protocols | |
|---|---|
| IF protocol for CL Plus 488 PQBP1 antibody CL488-84491-6 | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |