Epigenetics Modifications - DNA methylation
DNA Methylation: An Overview
Several types of epigenetic modifications
Chromatin structure can be modified by several epigenetic mechanisms, which can be divided into a number of categories: DNA methylation, covalent histone modifications, non-covalent mechanisms (histone variants and nucleosome remodeling), and non-coding RNAs (ncRNAs).
DNA methylation
DNA methylation is the most widely studied epigenetic mechanism. In eukaryotes, DNA methylation consists of the addition of a methyl group at the cytosines followed by guanines (CpG dinucleotides). CpGs are underrepresented in the genome and show a tendency to concentrate at specific clusters, called CpG islands. DNA methylation is usually a repressive mark in the tissue-specific gene silencing and allele-specific inactivation of the X-chromosome, which is associated with hypermethylation of CpG islands (1). However, recent data have questioned this assumption, and it is likely to be more complex.
Usually, only a very small amount of CpG island promoters are methylated in normal cells. When they are hypermethylated, it is usually in the context of tumorigenesis or abnormal development. About 70% of annotated genes, as well as housekeeping genes, tissue-specific, and regulatory genes, are related to CpG islands in their promoter regions (2), emphasizing the role of CpG islands in gene regulation (Figure 1). The current explanation for this unexpected frequency is related to the maintenance of chromosomal stability, translocation prevention, and endoparasitic sequence silencing (3).
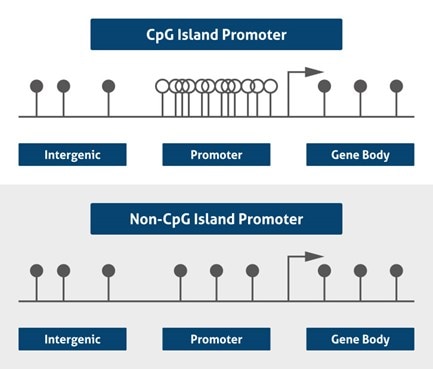 |
| Figure 1. CGI versus non-CGI promoter. Black dots represent methylated CpG and white dots are unmethylated (based on 4) |
The two methods of DNA methylation
There are two ways in which DNA methylation can control transcription by inhibition. The first is interference with the interaction of transcription factors and other DNA-binding proteins (5). The second method of control is the inhibition of gene expression by recruiting methyl-CpG binding protein (MBD) (6).
The enzymes that are involved in DNA methylation are called DNA methyltransferases (DNMTs). They catalyze the transfer of a methyl group from an S-adenosyl-L-methionine to cytosine residues. The mammalian DNMT family consists of five members: DNMT1, DNMT2, DNMT3A, DNMT3B, and DNMT3-like (DNMT3L) (7).
Only DNMT1, DNMT3A, and DNMT3B possess methyltransferase activity. The catalytic members of the DNMT family are classified into either de novo DNMT (DNMT3A and DNMT3B) or maintenance DNMT (DNMT1) groups. De novo DNMTs are highly expressed in stem cells and are downregulated after the process of differentiation. This modification consists of the addition of a methyl group at the cytosine residues of the DNA template. DNMTs enzymes catalyze either the de novo or maintenance methylation of hemimethylated.
DNA following DNA replication. They transfer a methyl group from the methyl donor S-adenosylmethionine (SAM), resulting in 5-methylcytosine (Figure 2). 5-methylcytosine is recognized by the methyl-CpG binding domain (MBD) of certain proteins. In mammals, there are five members: MeCP2, MBD1, MBD2, MBD3, and MBD4. Most of the MBD proteins are located in highly methylated chromatin regions, which influence silencing of imprinting genes and in endoparasitic sequences, promoting transcriptional repression and genomic stability (8).
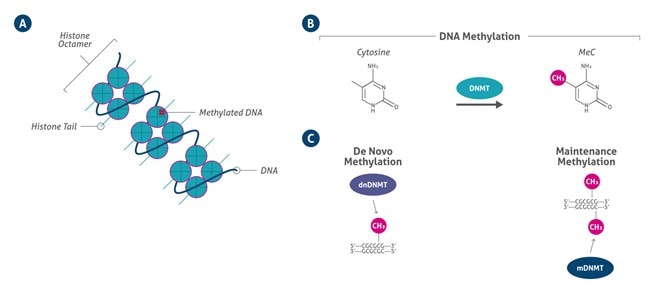 |
| Figure 2. A) DNA methylation can occur at cytosine bases when a methyl group is added at the 5′ position on the pyrimidine ring by a DNMT (B). De novo DNMTs methylate previously nonmethylated cytosines, whereas maintenance DNMTs methylate hemimethylated DNA at the complementary strand (C) (based on 9). |
DNA methylation is involved not only in the transcription process
DNA methylation is the oldest epigenetic mechanism known to correlate with gene expression (10). Besides regulating transcription process, DNA methylation has many other important biological functions. DNA methylation represses the expression of parasitic sequences, endogenous retrovirus, and transposable elements, such as α-satellites, a short stretch of DNA characterized by the action of the ALU restriction endonuclease – ALU-Yb8 and a long interspersed nuclear elements-1 (LINE-1).
Related Products
| Antibody | Cat No | Type | Applications | |
| DNMT1 | 24206-1-AP | Rabbit Poly | ELISA, IF, IP, WB | KD/KO Validated |
| DNMT3A | 19366-1-AP | Rabbit Poly | ELISA, WB, IHC | KD/KO Validated |
| DNMT3A | 20954-1-AP | Rabbit Poly | ELISA, IP, WB | |
| DNMT3L | 14939-1-AP | Rabbit Poly | ELISA, WB | |
| DNMT2 | 19221-1-AP | Rabbit Poly | ELISA, WB |
References
- Developmental programming of CpG island methylation profiles in the human genome.
- CpG islands and the regulation of transcription.
- DNA methylation patterns and epigenetic memory.
- Mining cancer methylomes: prospects and challenges.
- CpG islands and the regulation of transcription.
- DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein.
- The DNA methyltransferases of mammals.
- Proteins that bind methylated DNA and human cancer: reading the wrong words.
- DNA methylation and memory formation.
- DNA methylation and gene function.
Related Content
CRISPR-Cas9, TALENs and ZFNs - the battle in gene editing
How to solve the genome DNA damage
Epigenetics - DNA layers and chromatin modeling
Using Flow Cytometry with Epigenetics

Support
Newsletter Signup
Stay up-to-date with our latest news and events. New to Proteintech? Get 10% off your first order when you sign up.