Fluorescent protein tags
Types of fluorescent protein tags and comparisons
Introduction
Fluorescent proteins (FPs) have been used as protein tags since the mid-1990s mainly for cell biology and fluorescence microscopy. These tags have not only revolutionized cell biology by enabling the imaging of almost any protein, they are also used in biochemical applications. An important example is the immunoprecipitation and affinity purification of FP-tagged proteins, which was enabled by the development of affinity resins with high yield, purity, and affinity such as ChromoTek’s Nano-Traps.
In this blog we provide a review of
- green fluorescent proteins
- red fluorescent proteins
- self-labelling proteins, that require the covalent coupling of a fluorescent molecule
Types of fluorescent proteins
Most researchers use intrinsically fluorescent proteins GFP, mNeonGreen, TurboGFP, RFP, or mCherry. Alternatively, extrinsically fluorescent or self-labelling proteins have been introduced that require the covalent coupling of a fluorescent molecule to the non-fluorescent protein, e.g. the protein tags SNAP, CLIP, and Halo. These self-labelling fluorescent proteins have certain performance advantages over intrinsically FPs due to their fluorescent dyes’ properties.
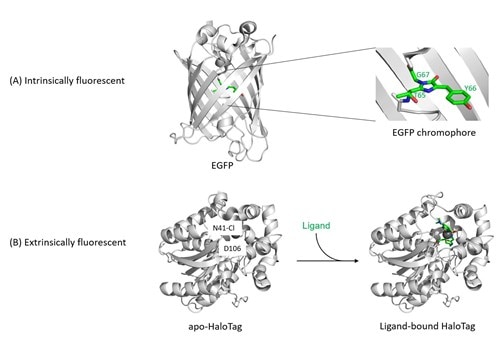
Figure 1: Structures of fluorescent proteins.
(A) Intrinsically fluorescent proteins (FPs) such as EGFP, GFP, RFP, mNeonGreen, turboGFP etc. share only a small number of common residues, but fold all into a conserved β-barrel structure. Their fluorescence arises through the backbone cyclization and oxidation of three amino acid residues in the center of this barrel (highlighted on the right), which results in a two-ring chromophore. This chemical process is described as maturation of the chromophore, is inherent to the protein fold, and depends only on environmental variables such as temperature and oxygen concentration, but not on additional enzymes. The colour, photostability, quantum yield, and other spectral properties of intrinsically fluorescent proteins are a result of mutations within the amino acids that make up the chromophore or that are located in the vicinity of the chromophore.
(B) Extrinsically fluorescent proteins such as HaloTag are non-fluorescent in their basal apo-form. Only if a suitable activated fluorophore is added to the HaloTag protein, this fluorophore will be captured and covalently bound by the HaloTag residue D106, turning HaloTag fluorescent. (PDB IDs for structures: EGFP, 2y0g; apo-HaloTag, 5uy1; holo-HaloTag, 5uxz.)
Green fluorescent proteins
Jellyfish Green Fluorescent Protein (GFP) and its derivatives are still the most frequently used fluorescent proteins in biomedical research. Recently, additional green fluorescent proteins have been introduced that are derived from other organisms. These FPs own the same basic fold as GFP but diverge widely on sequence-level. Therefore, they require novel, dedicated research tools such as antibodies.
The original: GFP
Green Fluorescent Protein was first isolated from the jellyfish Aequorea victoria in 1962 by Osamu Shimomura. It has a long Stokes shift green fluorescence (ex 395nm; em 509 nm). 30 years later, Douglas Prasher eventually managed to clone the sequence of GFP and Martin Chalfie expressed this sequence in vivo. Later, the Roger Tsien lab developed GFP into a suite of veritable research tools. Shimomura, Chalfie, and Tsien were awarded the Nobel Prize in 2008. See Roger Tsien’s Nobel Prize lecture here: https://www.nobelprize.org/prizes/chemistry/2008/tsien/lecture/.
Scientists developed a plethora of GFP variants with varying properties. These FPs have different functional and spectral properties. The first significant improvement of GFP was a mutation (S65T) that increased the intensity and stability of the fluorescence signal. The main excitation peak has been shifted to 488 nm (Heim et al., 1995). The common variant EGFP is an engineered version of GFP, which facilitates the practical use of GFP in a variety of different organisms and cells.
Green Fluorescent Protein GFP is also known as avGFP, wtGFP, and gfp10; EGFP as enhanced GFP and GFPmut1.
https://www.fpbase.org/protein/avgfp/
https://www.fpbase.org/protein/egfp/
TurboGFP
Reported in 2004, TurboGFP is a fast maturing and bright dimeric green fluorescent protein derived from CopGFP from the copepod Pontellina plumata. Copepod TurboGFP is evolutionarily distant from jellyfish-derived fluorescent proteins such as EGFP and shares only about 20% sequence identity with the commonly used GFP variants. Therefore, most anti-GFP antibodies including the GFP-Nanobody used in GFP-Trap do not bind to TurboGFP.
https://www.fpbase.org/protein/turbogfp/
mNeonGreen
mNeonGreen is derived from a multimeric yellow fluorescence protein of the lancelet Branchiostoma lanceolatum. Hence, mNeonGreen is evolutionarily distant from jellyfish-derived FPs. mNeonGreen and common GFP derivatives share just about 20% sequence identity. Due to the low sequence similarity, it is expected that affinity tools (i.e. antibodies) for GFP variants will not bind to mNeonGreen. Certainly, this has been shown for ChromoTek’s affinity reagents (anti-GFP Nanobodies/ VHHs and antibodies).
First published in 2013, mNeonGreen is up to three times brighter than GFP. It is an emerging, monomeric versatile green/yellow fluorescent protein for imaging applications including super- resolution microscopy. Furthermore, mNeonGreen acts as acceptor for cyan fluorescent proteins in fluorescence resonance energy transfer (FRET) applications. It seems to be the brightest monomeric green fluorescent protein known so far and has a fast maturation rate.
https://www.fpbase.org/protein/mneongreen/
Comparison of GFP, TurboGFP, and mNeonGreen
Property |
EGFP (most commonly used GFP derivative) |
turboGFP |
mNeonGreen |
|---|---|---|---|
|
Discovery/ first publication |
1995 |
2004 |
2013 |
|
Origin |
GFP from Jellyfish |
CopGFP from copepod Pontellina plumata |
multimeric yellow fluorescent protein from lancelet Branchiostoma lanceolatum |
|
Maturation rate (at 37°C) |
25 min |
25 min |
10 min |
|
Sequence identity with EGFP |
(100%) |
~20% |
~20% |
|
Derived from GFP? |
Yes |
No |
No |
|
Structure |
Weak dimer |
Dimer |
Monomer |
|
Common variants |
AcGFP, Clover, eGFP, Emerald, GFP, GFP5, GFP Envy, GFP S65T, mGFP, mPhluorin, PA-GFP, Superfolder GFP, TagGFP, TagGFP2, monomeric eGFP A206K, CFP, eCFP, mCerulean, YFP, Citrine, eCitrine, eYFP, Venus, Ypet, BFP |
TurboGFP, CopGFP |
mNeonGreen |
|
Excitation/ Emission maximum |
488 nm/ 509 nm |
482 nm/ 502 nm |
506 nm/ 517 nm |
|
Length/ molecular weight (MW) |
239 amino acids, |
2x 232 amino acids, |
237 amino acids, |
|
Research tools offered by ChromoTek |
Immunoprecipitation: GFP-Trap Western blot: Rat anti-GFP antibody [3H9] Control: purified EGFP protein |
Immunoprecipitation: TurboGFP-Trap |
Immunoprecipitation: mNeonGreen-Trap Western blot: Mouse anti-mNeonGreen antibody [32F6]
|
Red fluorescent proteins
Red fluorescent proteins (RFPs) are FPs that emit red-orange fluorescence light. The first RFP that became commercially available was DsRed. It was derived from Discosoma sp. sea anemones in 1999.
DsRed has some immanent practical problems: (i) It has a maturation time of about 24 hours, which makes it unusable for short(er) time experiments. (ii) The tetrameric form of DsRed may compromise the function of proteins to which it is attached. (iii) Its photostability is rather low.
In consequence, DsRed was subjected to site-directed mutagenesis to become commonly applicable as a genetically encoded fusion tag. Eventually, monomeric RFP derivatives with better fluorescent performance (in terms of brightness and photostability) and higher maturation efficiency were created. In addition, derivatives were generated with orange, red, and far red fluorescence. These monomerized versions of RFP became the valuable research tools mCherry, mOrange, mRaspberry, mPlum (also known as the “mFruits”), mKO, mRFP (a.k.a. mRFP1), mRFPruby, mRuby, tagRFP, mKate2, and DsRed-Express etc.
Additional RFPs were identified in other anthozoans (i.e. anemones and corals), but these proteins were mostly tetramers, too. Thus, they have not been further optimized for use in research yet.
mRFP (also known as mRFP1)
The first monomeric variant of dsRed, genetically engineered in Roger Tsien’s lab, was simply designated mRFP (or mRFP1), i.e. monomeric red fluorescent protein. Compared to dsRed, mRFP1 is characterized by slightly lower levels of absorption, quantum yield, and photostability. However, its maturation rate is about 10 times faster than that of DsRed, which results in a similar effective brightness when expressed in living cells.
https://www.fpbase.org/protein/mrfp1/
mCherry
mCherry is probably the most commonly used RFP variant. It is a monomeric red fluorescent protein with broad applicability as a fusion protein in various cell types. Like other mFruit RFPs, mCherry is derived from the dsRed variant mRFP1 via directed evolution by Roger Tsien’s lab. Compared to other mFruits, mCherry has the highest photostability, fastest maturation rate, and excellent pH resistance. It has, however, a lower quantum yield than mRFP1.
https://www.fpbase.org/protein/mcherry/
mPlum
Roger Tsien’s lab has also generated a far-red monomeric derivative of mRFP1/dsRed, named mPlum. Far-red FPs are beneficial for whole-body imaging applications because the main tissue absorbers such as water, lipids and hemoglobin are nearly transparent at the emission range of 650-900 nm. Like most red-shifted RFPs, mPlum features an extended Stokes shift.
https://www.fpbase.org/protein/mplum/
Comparison of mCherry, mRFP (mRFP1), and mPlum
Property |
mCherry |
mRFP (mFRP1) |
mPlum |
|---|---|---|---|
|
Discovery/ first publication |
2004 |
2002 |
2004 |
|
Origin |
DsRed from sea anemone |
DsRed from sea anemone |
DsRed from sea anemone |
|
Maturation rate (at 37°C) |
15 min |
60 min |
100 min |
|
Structure |
Monomer |
Monomer |
Monomer |
|
Aggregation |
no |
no |
no |
|
Excitation/ Emission maximum |
587 nm/ 610 nm |
584 nm/ 607 nm |
590 nm/ 649 nm |
|
Length/ molecular weight (MW) |
236 amino acids, |
228 amino acids, |
229 amino acids, |
|
Research tools provided by ChromoTek |
Immunoprecipitation: RFP-Trap |
Immunoprecipitation: RFP-Trap |
Immunoprecipitation: RFP-Trap |
Halo, SNAP, and CLIP
The extrinsically fluorescent protein tags Halo, SNAP, and CLIP require the covalent capture of a small fluorescent ligand to turn into a fluorescent protein. To this end, these self-labelling protein tags are derived from enzymes that catalyze the formation of chemical bonds: The SNAP and CLIP tags are variants of O6-alkylguanine-DNA alkyltransferase that react with benzylguanine and benzylcytosine derivatives, respectively. The HaloTag is derived from a haloalkane dehalogenase and reacts with alkylhalides (Figure 1).
In general, both intrinsically and extrinsically fluorescent proteins are fused to a protein of interest to enable its cellular imaging and detection. However, extrinsically fluorescent proteins require the addition of a reactive fluorophore, which has several advantages:
- Higher quantum yield and photostability
- Strong fluorescence in both live and fixed cells
- A broader selection of fluorescent dyes
- A single genetic construct allows the choice of distinct fluorophores for multicolor imaging/multiplexing
- Fluorescence only initiated upon addition of the label
The availability of three extrinsically fluorescent proteins with different substrate/ligand specificities enables their orthogonal use in multiplexed experiments, also in combination with intrinsically FPs.
Depending on the experimental needs, one may use cell permeant ligands based on tetramethylrhodamine (TMR), Oregon Green, diAcFAM, or Coumarin, which readily cross the cell membrane for labeling intracellular proteins. Alternatively, cell impermeant ligands based on impermeable fluorophores like Alexa Fluor® 488 and 660 may be applied for quick cell surface labeling.
Comparison of HaloTag, SNAP-Tag, and CLIP-Tag
Property |
HaloTag |
SNAP-Tag |
CLIP-Tag |
|---|---|---|---|
|
Discovery/ first publication |
2008 |
2008 |
2008 |
|
Origin |
Haloalkane dehalogenase from Rhodococcus rhodochrous |
human O6-alkylguanine-DNA-alkyltransferase |
human O6-alkylguanine-DNA-alkyltransferase |
|
Structure |
Monomer |
Monomer |
Monomer |
|
Reactivity |
Chloroalkane derivatives |
O6-benzylguanine derivatives |
Benzylcytosine derivatives |
|
Ligands |
Cell permeable or non-permeable dyes, fluorophores, biotin, and beads |
Cell permeable or non-permeable dyes, fluorophores, biotin, and beads |
Cell permeable or non-permeable dyes, fluorophores, biotin, and beads |
|
Excitation/ Emission maximum |
Depends on coupled fluorophore |
Depends on coupled fluorophore |
Depends on coupled fluorophore |
|
Length/ molecular weight (MW) |
297 amino acids, |
182 amino acids, |
182 amino acids, |
|
Research tools offered by ChromoTek |
Immunoprecipitation: Halo-Trap |
Immunoprecipitation: SNAP/CLIP-tag-Trap |
Immunoprecipitation: SNAP/CLIP-tag-Trap |
HaloTag
The self-labelling HaloTag has been derived from the haloalkane dehalogenase enzyme DhaA from Rhodococcus rhodochrous. Its active site has been genetically modified to irreversibly bind chloroalkane linker substrates. Owing to this type of suicide inhibition, regeneration of its catalytic site for further dehalogenation is not possible anymore. Depending on the substrate chosen, the HaloTag converts into a fluorescent protein tag or may be immobilized on agarose beads, for example.
As haloalkane dehalogenases are absent in eukaryotic cells and most prokaryotes, including E. coli, there will be no background labelling.
SNAP-tag
The self-labeling protein tag SNAP-tag is derived from the human O6-alkylguanine-DNA-alkyltransferase (hATG), which, as a wildtype protein, removes alkylation damage from DNA. The resulting hAGT variant used as SNAP-tag reacts covalently with O6-benzylguanine derivatives (i.e. a fluorescent label conjugated to guanine or chloropyrimidine leaving groups via a benzyl linker) in an irreversible and highly specific manner. In the labeling reaction, the substituted benzyl group of the substrate is covalently attached to the SNAP-tag.
CLIP-tag
CLIP-tag is a modified version of SNAP-tag, engineered to react with benzylcytosine rather than benzylguanine derivatives. When used in conjunction with SNAP-tag, CLIP-tag enables the orthogonal and complementary labeling of two proteins simultaneously in the same cell.
References
A guide to choosing fluorescent proteins
Shaner N.C., Steinbach P.A. and Tsien R.Y. (2005)
Nature Methods 2(12), 905-909 doi: 10.1038/nmeth819
http://www.tsienlab.ucsd.edu/Publications/Shaner%202005%20Nature%20Methods%20-%20Choosing%20fluorescent%20proteins.pdf
Green Fluorescent Proteins:
The green fluorescent protein
Tsien R.Y. (1998)
Annual Review of Biochemistry, 67(1), 509-544. doi: 10.1146/annurev.biochem.67.1.509.
Improved green fluorescence
Heim, R., Cubitt A.B. and Tsien R.Y. (1995)
Nature, 373, 663-664. doi: 10.1038/373663b0
Structural basis for the fast maturation of Arthropoda green fluorescent protein
Evdokimov A.G., Pokross M.E., Egorov N.S., Zaraisky A.G., Yampolsky I.V., Merzlyak E.M., Shkoporov A.N., Sander I, Lukyanov K.A and Chudakov D.M. (2006)
EMBO reports, 7(10), 1006-1012. doi: 10.1038/sj.embor.7400787.
A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum
Shaner N.C., Lambert G.G., Chammas A., Ni Y., Cranfill P.J. Baird M.A., Sell B.R., Allen J.R., Day R.N., Israelsson M, Davidson M.W. and Wang J. (2013)
Nature Methods, 10(5), 407-409. doi: 10.1038/nmeth.2413
Red Fluorescent Proteins:
Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein
Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N.G., Palmer A.E. and Tsien R.Y. (2004)
Nature Biotechnology, 22(12), 1567-1572. doi: 10.1038/nbt1037
A monomeric red fluorescent protein
Campbell R.E., Tour O., Palmer A.E., Steinbach P.A., Baird G.S., Zacharias D.A. and Tsien R.Y. (2002)
PNAS, 99(12), 7877-7882. doi: 10.1073/pnas.082243699
Evolution of new nonantibody proteins via iterative somatic hypermutation
Wang L., Jackson W.C., Steinbach P.A. and Tsien R.Y. (2004)
PNAS, 101(48), 16745-16749. doi: 10.1073/pnas.0407752101.
Recovery of Red Fluorescent Protein Chromophore Maturation Deficiency through Rational Design
Moore M.M., Oteng-Pabi S.K., Pandelieva A.T., Mayo S.L. and Chica R.A. (2012)
Plos ONE, 7(12), e52463. doi: 10.1371/journal.pone.0052463.
Halo, SNAP, and CLIP:
Releasable SNAP-tag Probes for Studying Endocytosis and Recycling
Cole N.B. and Donaldson J.G. (2012)
ACS Chemical Biology,7(3): 464-469, doi: 10.1021/cb2004252
Site-Specific Protein Labeling with SNAP-Tags
Cole N.B. (2013)
Current protocols in protein science / editorial board, Coligan J.E. et al., 73: 30.1.1-30.1.16., doi: 10.1002/0471140864.ps3001s73
An engineered protein tag for multiprotein labeling in living cells
Gautier A., Juillerat A., Heinis C., Corrêa I.R., Kindermann M., Beaufils F. and Johnsson K. (2008)
Chemistry and Biology 15 (2): 128–136, doi: 10.1016/j.chembiol.2008.01.007
HaloTag: A Novel Protein Labeling Technology for Cell Imaging and Protein Analysis
Los G.V., Encell L.P., McDougall M.G., Hartzell D.D., Karassina N., Zimprich C, Wood M.G., Learish R, Friedman-Ohana R, Urh M., Simpson D., Mendez J., Zimmerman K., Otto P., Vidugiris G., Zhu J., Darzins A., Klaubert D.H., Bulleit R.F. and Wood K.V. (2008)
ACS Chem. Biol. 3(6): 373–382 doi: 10.1021/cb800025k
A general method for the covalent labeling of fusion proteins with small molecules in vivo.
Keppler A., Gendreizig S., Gronemeyer T., Pick H., Vogel H. and Johnsson K (2003)
Nat. Biotechnol. 21: 86–89, doi: 10.1038/nbt765
Fluorescent Labeling of COS-7 Expressing SNAP-tag Fusion Proteins for Live Cell Imaging
Provost C.R. and Sun L. (2010)
Visualized Exp. 39: 1876, doi: 10.3791/1876
For research use only.
ChromoTek’s products and technology are covered by granted patents and pending applications owned by or available to ChromoTek GmbH by exclusive license.
ChromoTek, Chromobody, F2H, GFP-Trap, Myc-Trap, RFP-Trap, Spot-Tag, Spot-Label, and Spot-Trap are registered trademarks of ChromoTek GmbH. SNAP-tag is a registered trademark and CLIP-tag is a trademark of New England Biolabs, Inc. Nanobody is a registered trademark of Ablynx, a Sanofi company. Other suppliers’ products may be trademarks or registered trademarks of the corresponding supplier each. Statements on other suppliers’ products are given according to our best knowledge.
Related Content
Why are recombinant Nanobodies/ VHHs beneficial?
Green fluorescent protein (GFP) in plant research
The best anti-GFP antibody for immunoprecipitation: GFP-Trap
What GFP-Trap should I use for my immunoprecipitation?
TurboGFP: Properties, Sequence, MW, origin
Mass spec-compatible immunoprecipiation for GFP, mNeonGreen, Myc, RFP, Spot, and TurboGFP
GFP Nanobody for better images in immunofluorescence
GFP and RFP-Booster for better immunofluorescence imaging
New anti-mNeonGreen antibody & Nano-Trap for Immunofluorescence & Immunoprecipitation

Support
Newsletter Signup
Stay up-to-date with our latest news and events. New to Proteintech? Get 10% off your first order when you sign up.