Mitochondria Antibodies
The center of cellular signaling and energetic balance
|
Mitochondrial Respiratory Complexes |
Introduction
Mitochondria are important cellular organelles that maintain cellular energy balance, contain key regulators of cell death processes, and play a significant role in cellular oxidative stress generation and maintenance of calcium homeostasis.
Links to cancer, apoptosis, autophagy, and hypoxia have brought mitochondria to the forefront of scientific studies in recent years. Knowledge of the subcellular location of a protein may reveal the potential role it plays in a variety of cellular processes.
Proteintech offers approximately all the antibodies needed for mitochondria research.
Blog post:Mitochondria: Center of cellular signaling and energetic balance |
Mitochondria Research Slideshow |
Mitochondrial markers
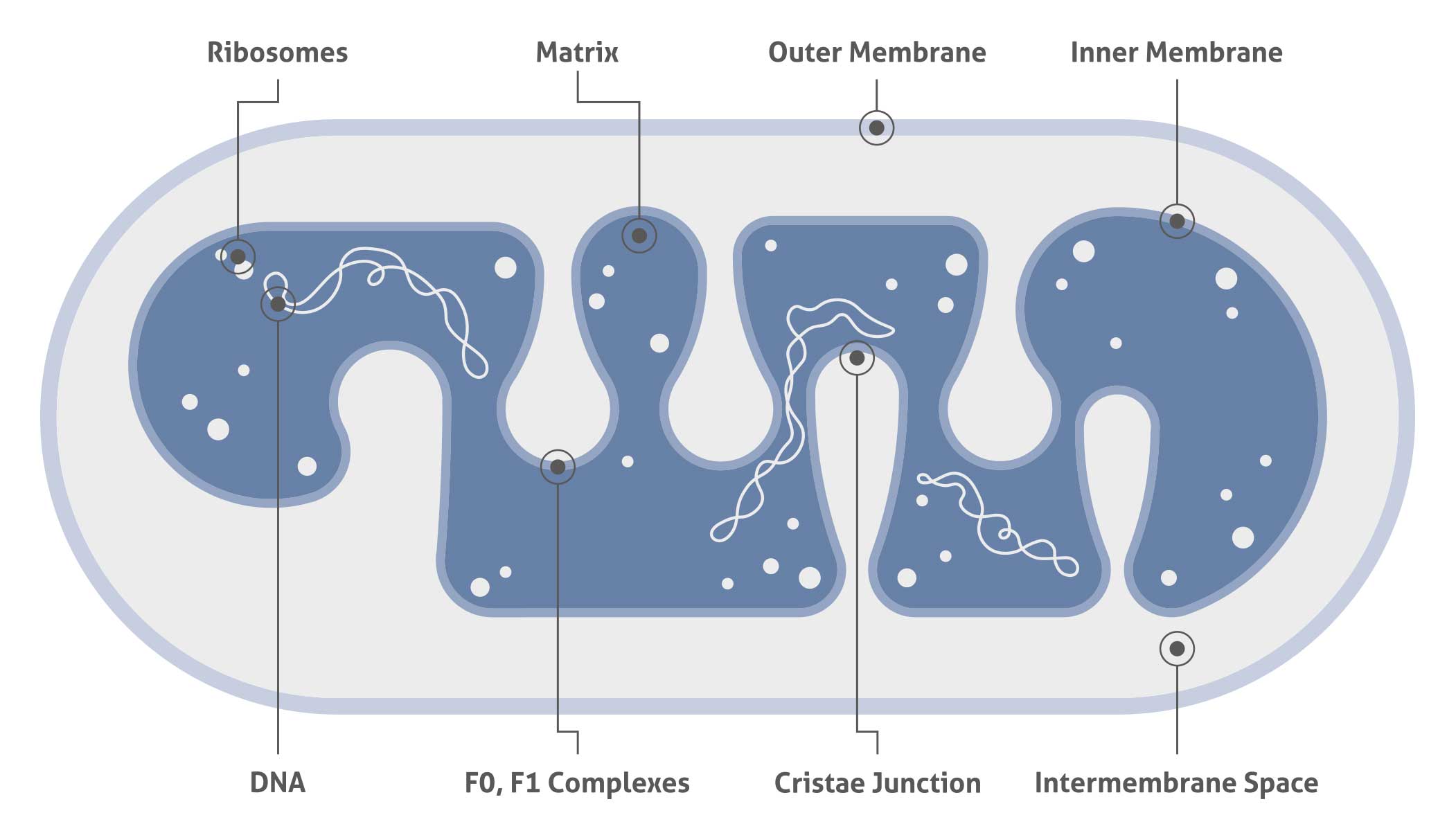
Mitochondria are composed of the inner and outer membranes, the intermembrane space, the cristae, and the matrix, and they contain their own DNA separated from the nucleus. Knowledge of the subcellular location of a protein may reveal the potential role it plays in a variety of cellular processes. Colocalization with one of the organelle-specific markers listed here can confirm the subcellular location of a mitochondrial protein of interest.
| Antibody Name | |||
| AIF | COX7A2L | HADHA | SLC25A20 |
| AK2 | COXIV | HSP60 | SMCR7L |
| ALDH1B1 | COXIV | MFF | SURF1 |
| ATP5A1 | CPT1A | MFN2 | SYNJ2BP |
| ATP5F1 | ECH1 | Mitofilin | TOM20 |
| ATP5H | ECHS1 | NDUFV2 | TOM40 |
| BCS1L | ETFA | NLRX1 | UQCRC1 |
| CLPP | F1S1 | ATP50 | |
| COX2 | GCSH | OXCT1 | |
| COX5B | GLUD2 | PMPCB |
| HSPD1 |
| Mitochondrial matrix and inner mitochondrial membrane marker |
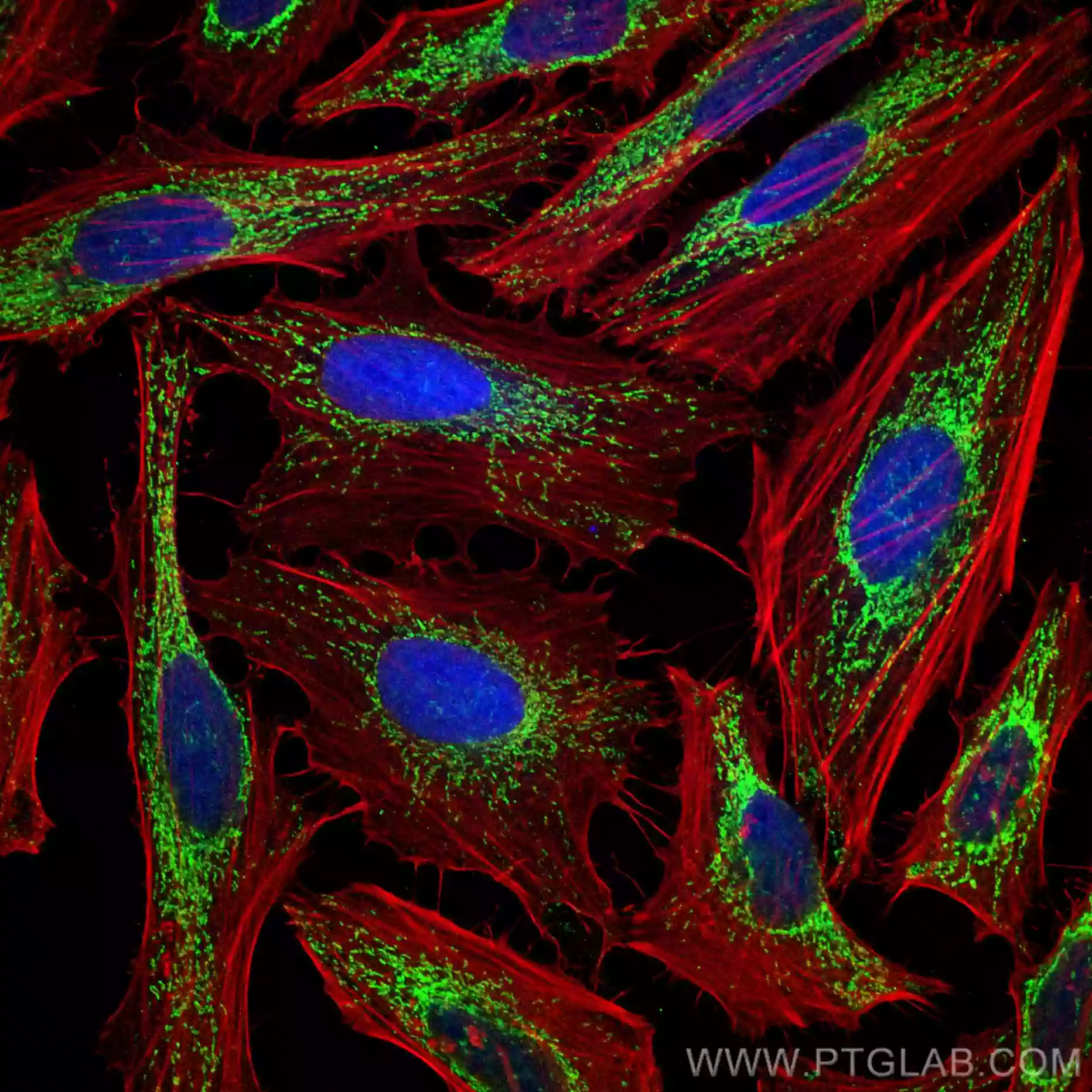 |
| Immunofluorescent analysis of (4% PFA) fixed HeLa cells using HSP60 antibody (66041-1-Ig, Clone: 2F10E7 ) at dilution of 1:100 and CoraLite®488-Conjugated AffiniPure Goat Anti-Mouse IgG(H+L), (CL594-Phalloidin, red).. |
| SCOT |
| Mitochondrial matrix marker |
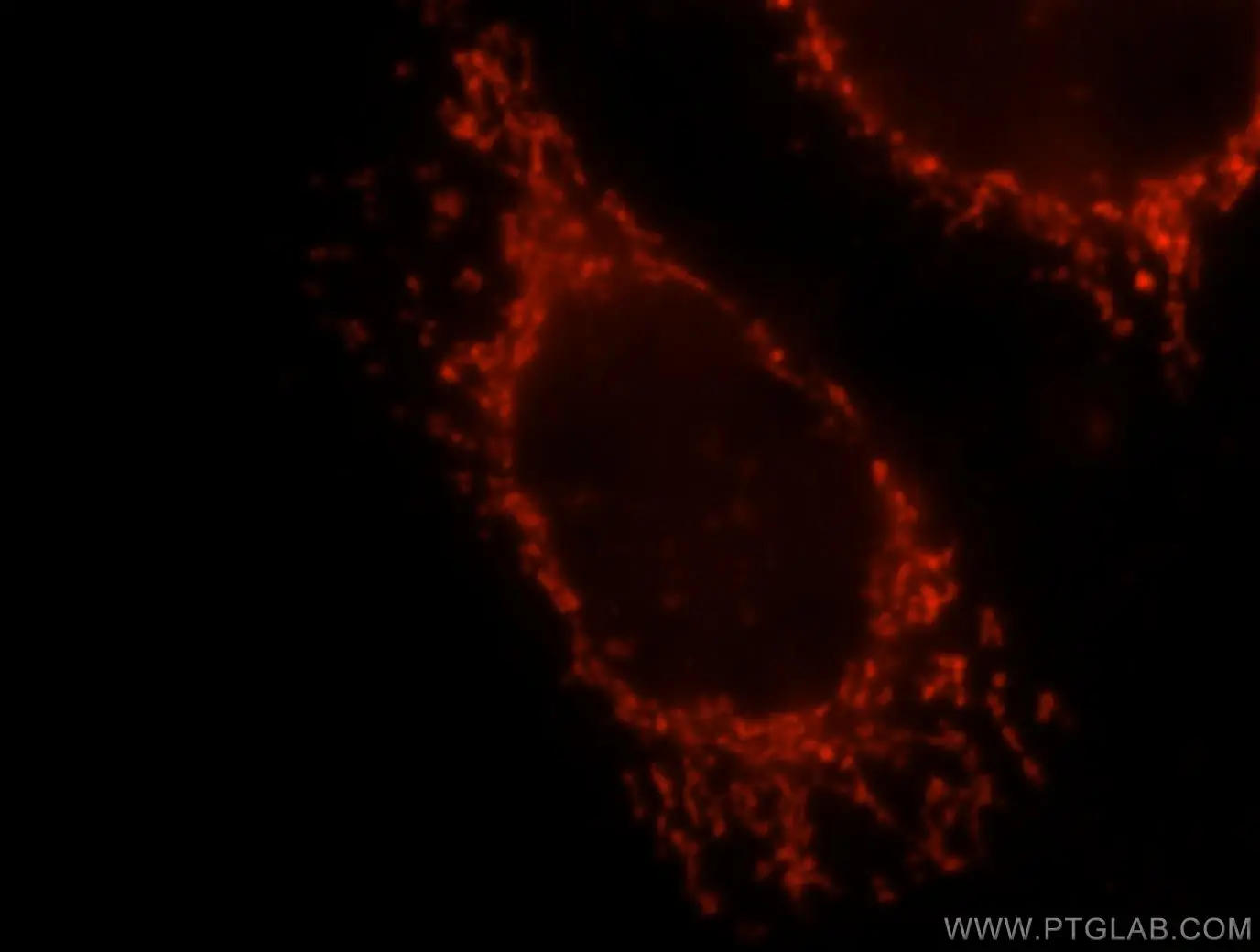 |
| Immunofluorescent analysis of MCF-7 cells, using OXCT1 antibody 12175-1-AP at 1:25 dilution and Rhodamine-labeled goat anti-rabbit IgG (red).. |
| AIFM1 |
| Mitochondrial inter-membrane space marker |
 |
| Immunofluorescent analysis of (4% PFA) fixed HeLa cells using AIF antibody (17984-1-AP) at dilution of 1:200 and CoraLite®488-Conjugated AffiniPure Goat Anti-Rabbit IgG(H+L), CL594-phalloidin (red). |
| TOMM40 |
| Mitochondrial outer membrane marker |
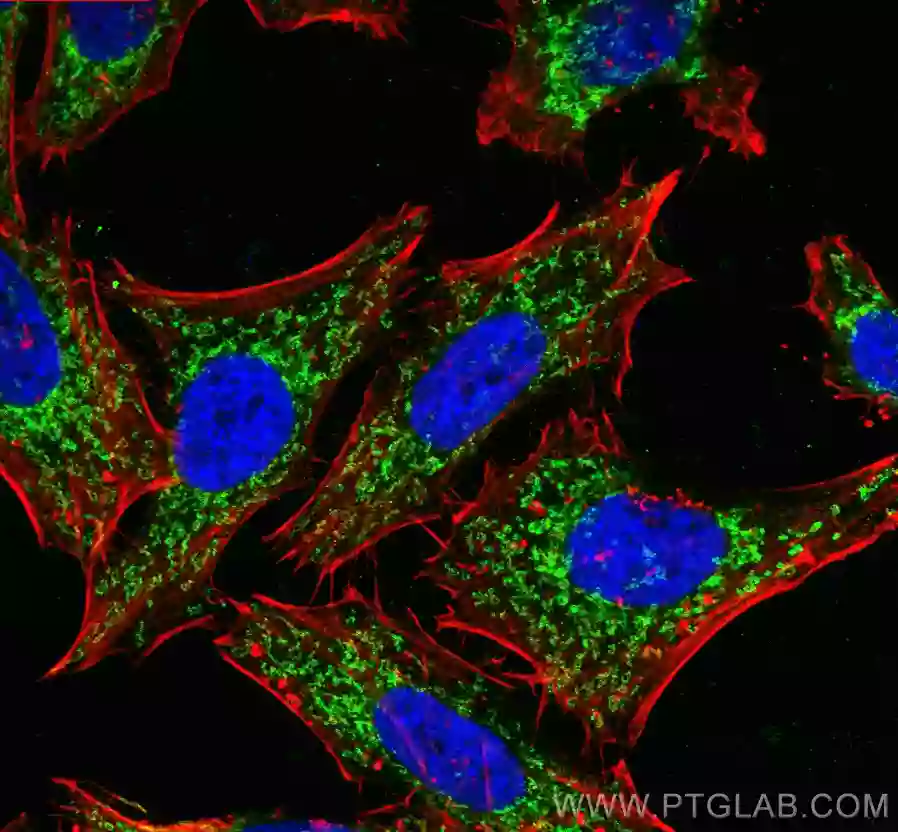 |
| Immunofluorescent analysis of (4% PFA) fixed HepG2 cells using TOMM40 antibody (18409-1-AP) at dilution of 1:400 and CoraLite®488-Conjugated AffiniPure Goat Anti-Rabbit IgG(H+L), (CL594-Phalloidin, red). |
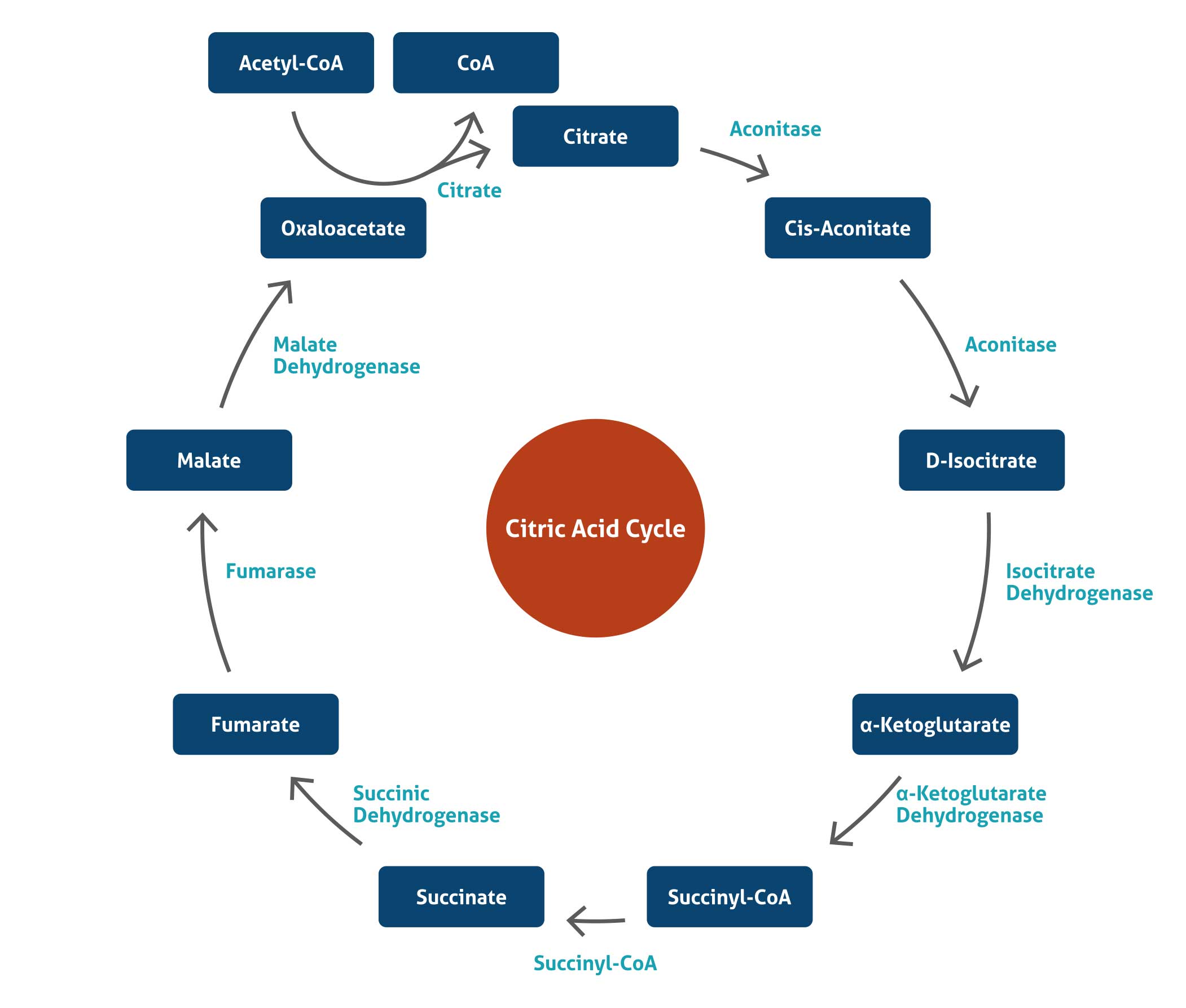
Citric Acid Cycle
The citric acid cycle – also known as the tricarboxylic acid (TCA) cycle or the Krebs cycle – is a series of chemical reactions used by all aerobic organisms to generate energy through the oxidation of acetate derived from carbohydrates, fats, and proteins into carbon dioxide and chemical energy in the form of adenosine triphosphate (ATP). In addition, the cycle provides precursors of certain amino acids as well as the reducing agent NADH that is used in numerous other biochemical reactions. In eukaryotic cells, the citric acid cycle occurs in the matrix of the mitochondrion. The reactions of the cycle are carried out by 8 enzymes that completely oxidize acetate, in the form of acetyl-CoA, into water and two molecules of carbon dioxide.
Related antibodies
| Antibody Name | ||
| Aconitase 2 | IDH3A | |
| Citrate synthase | MDH1 | |
| DLD | OGDH | |
| Fumarase | OGDHL | |
| IDH2 | SdhA | |
Blog post:Mitochondria are more than the powerhouse of the cell |
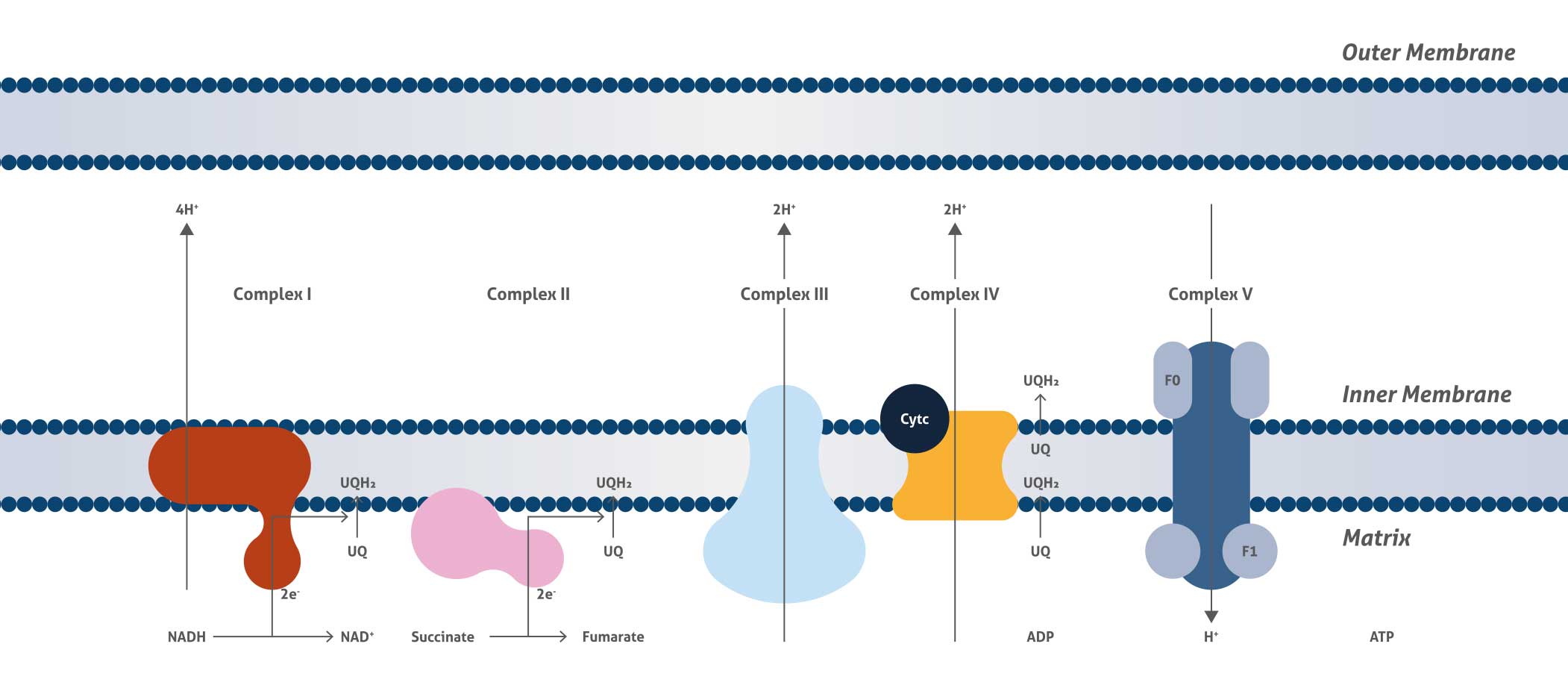
Mitochondrial Respiratory Complexes
Cellular respiration is the process that releases energy from food and supplies energy for life processes. The mitochondrial respiratory chain is the final and most important step for cellular respiration and is located on the inner membrane of the mitochondrion and comprises four large trans-membrane protein complexes (respiratory chain Complexes I, II, III, and IV) (CI, CII, CIII, CIV) as well as ubiquinone between CI/II and III and cytochrome c between CIII and IV. The function of the mitochondrial respiratory chain is biological oxidation by transferring electrons from NADH and succinate to oxygen and then generating proton gradient across the inner membrane.
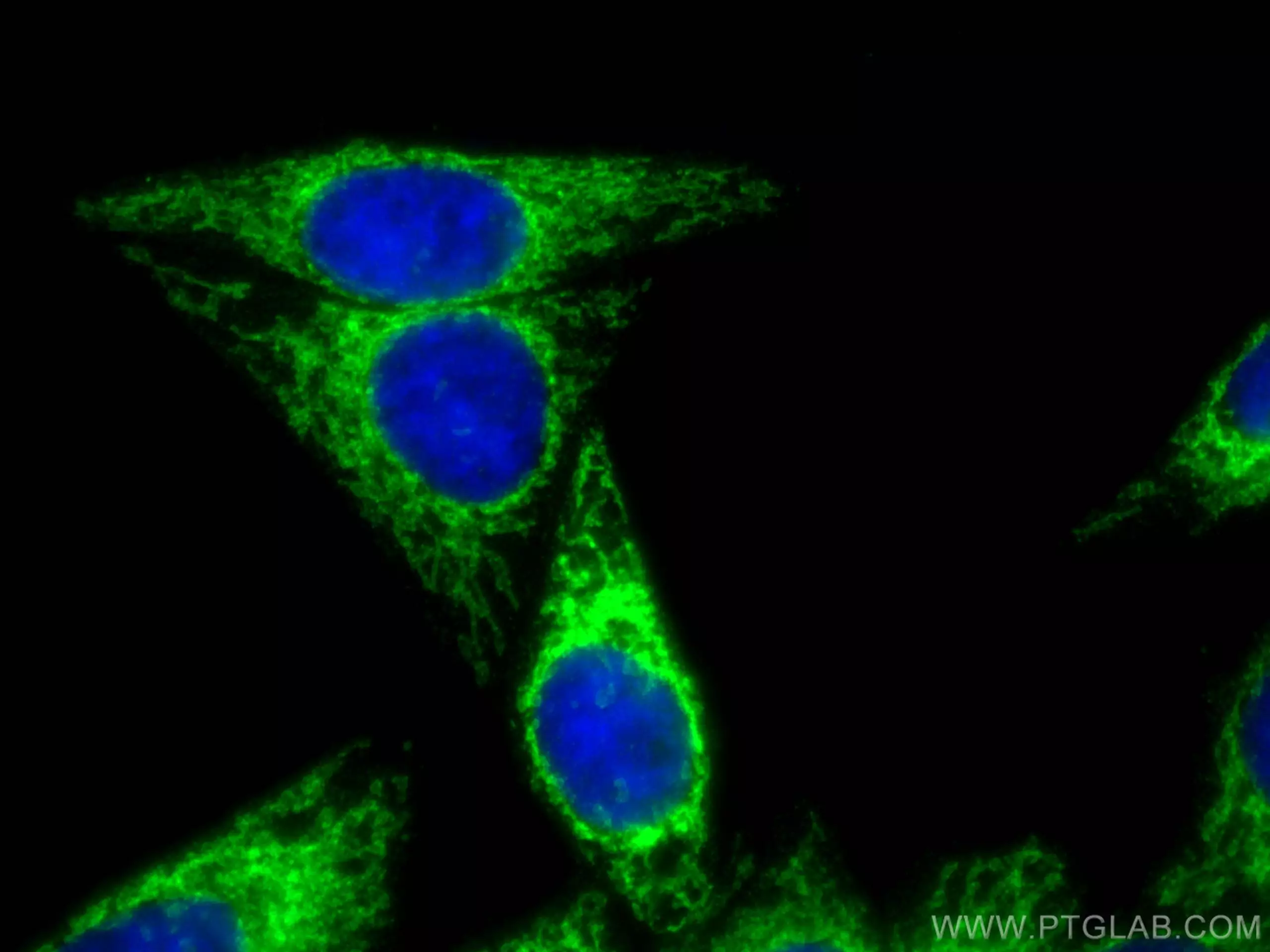
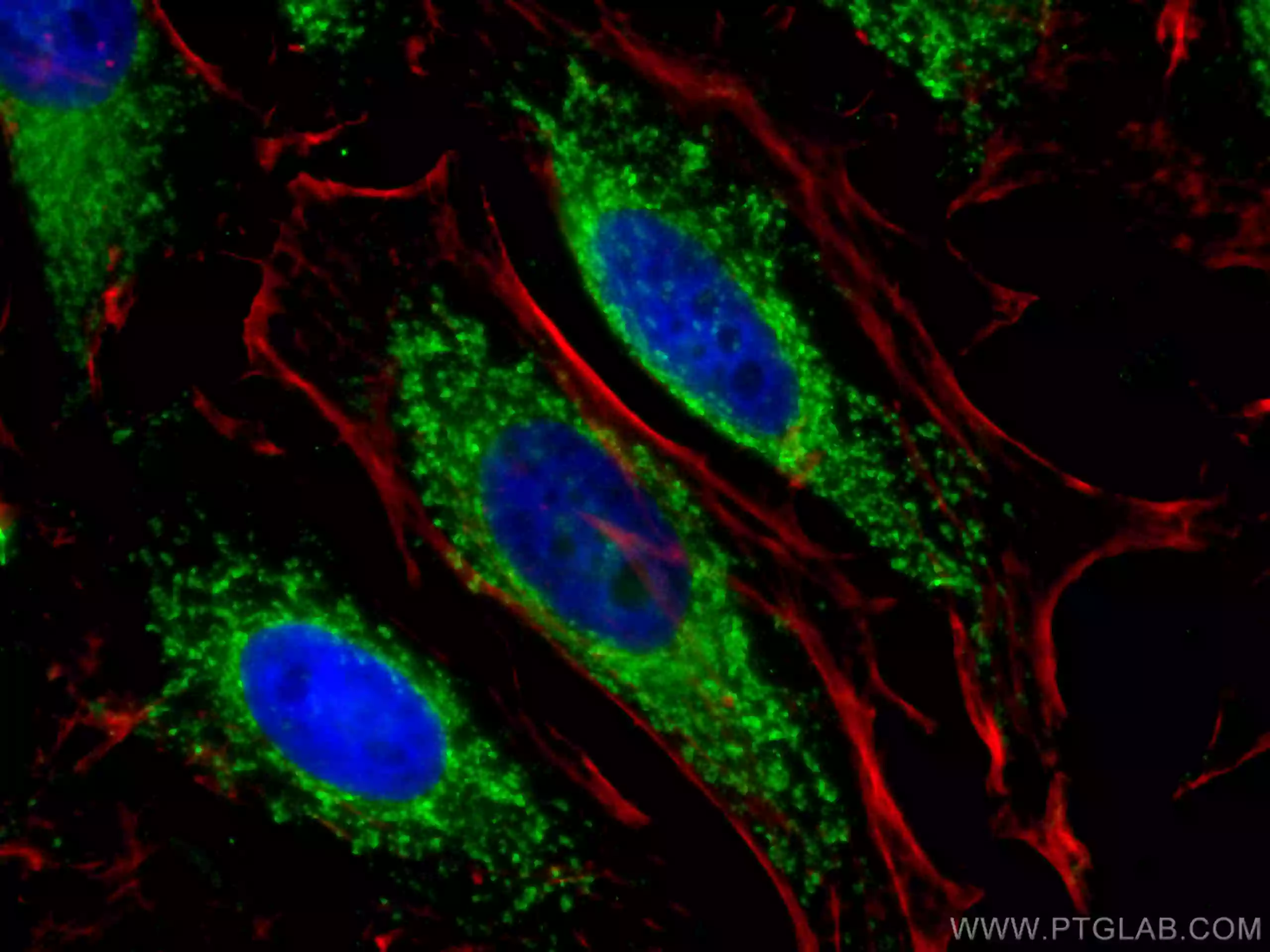
Mitochondrial Fission
Mitochondria are integral to cellular function and are responsible for energy production in eukaryotes, synthesis of metabolites, phospholipids, and heme, and maintenance of calcium homeostasis. Mitochondria are remarkably dynamic organelles undergoing frequent fusion and fission events. The opposing processes of fission and fusion maintain mitochondrial morphology and it is this equilibrium that ensures maintenance of mtDNA and metabolic mixing, bioenergetic functionality, and organelle number.
| MFF | SMCR7L |
|
Immunofluorescent analysis of (4% PFA) fixed HepG2 cells using MFF antibody (17090-1-AP) at dilution of 1:200 and CoraLite®488-Conjugated AffiniPure Goat Anti-Rabbit IgG(H+L). |
Immunofluorescent analysis of (4% PFA) fixed mouse testis tissue using SMCR7L antibody (67808-1-Ig, Clone: 3B3G3 ) at dilution of 1:200 and CoraLite®488-Conjugated AffiniPure Goat Anti-Mouse IgG(H+L). DAPI (blue). |
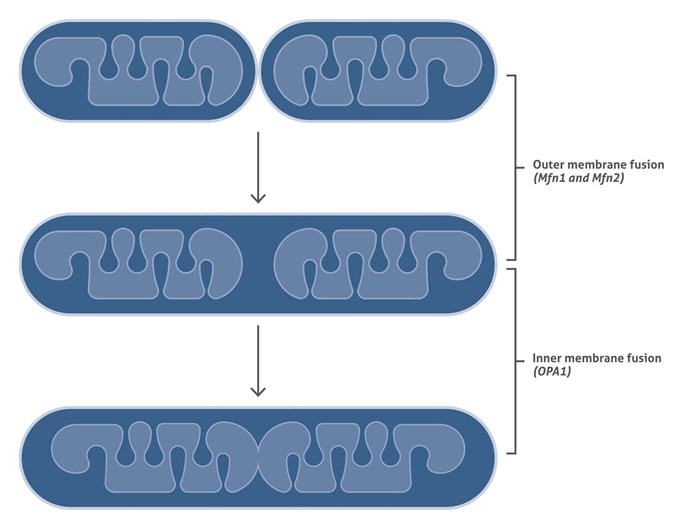
Mitochondrial Fusion
Mitochondrial fusion occurs in three stages:
1. Docking of two mitochondria via their outer membranes.
2. Fusion of outer membranes
3. Fusion of inner membranes.
The first two stages are mediated by the mitofusion proteins: Mfn1 and Mfn2 in mammals. The next stage – fusion of the inner mitochondrial membranes – is mediated by the dynamin-related GTPase OPA1
Mitochondrial Mediated Apoptosis
Apoptosis is the process of programmed cell death (PCD) that plays a central role in animal development and tissue homeostasis. There are two major apoptotic pathways known to date, initiated by either the mitochondria (the ‘intrinsic’ pathway) or the cell surface receptors (the ‘extrinsic’ pathway). Mitochondria-mediated apoptosis occurs in response to a wide range of death stimuli, including activation of tumor suppressor proteins (such as p53) and oncogenes (such as c-Myc), DNA damage, chemotherapeutic agents, serum starvation, and ultraviolet radiation.
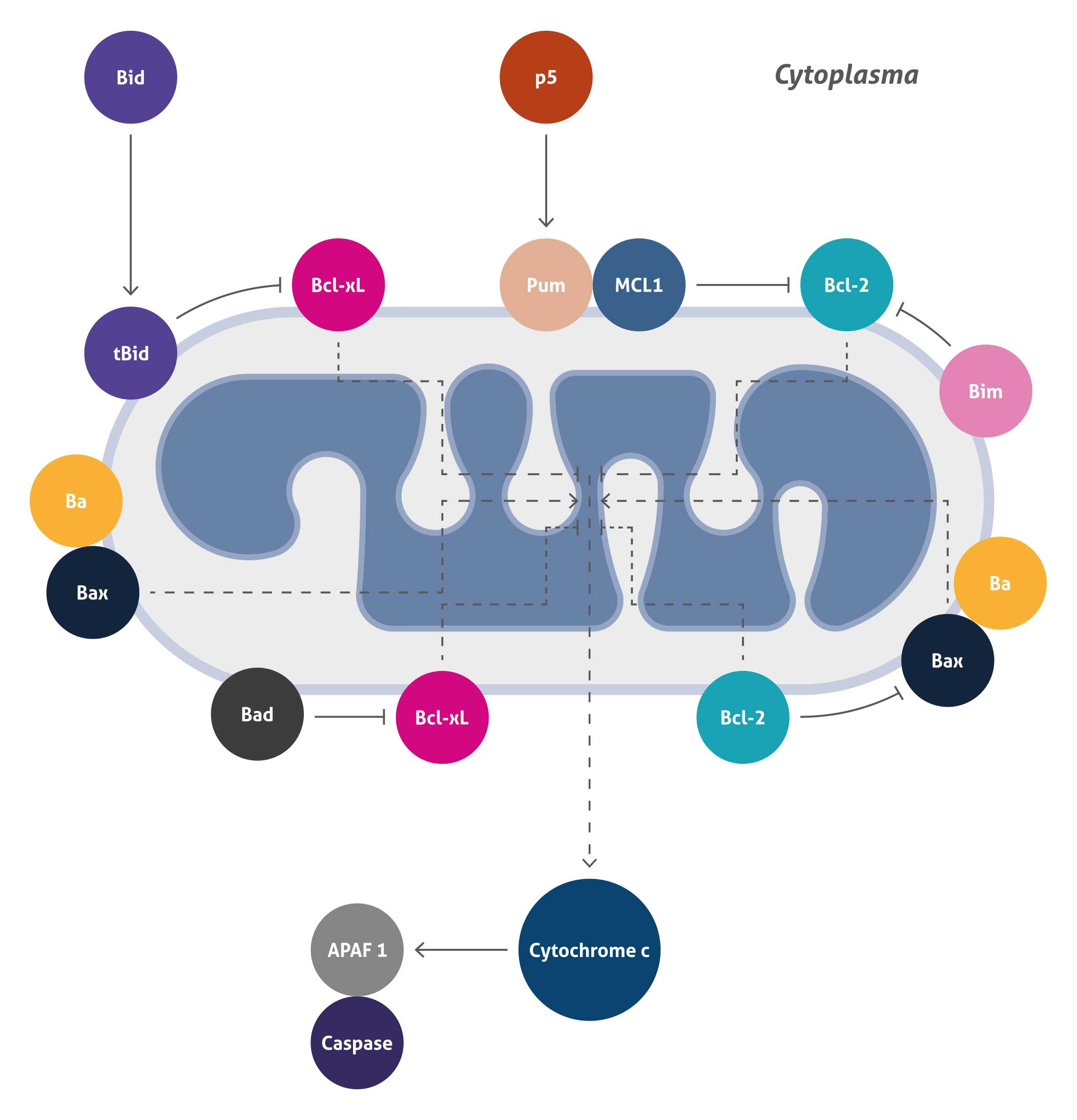
This pathway is initiated within the cell and results in increased mitochondrial permeability leading to release of pro-apoptotic molecules Cytochrome c into the cytoplasm.
Mitochondrial Autophagy
Mitochondrial autophagy is the process of selective removal of damaged mitochondria by autophagosomes and subsequent catabolism by lysosomes. One common mechanism is that mitochondrial depolarization results in PINK1 stabilization and Parkin recruitment to the mitochondria. Parkin can ubiquitinate Mitofusins 1 and 2 (MFN1 and 2), hexokinases, TOM complex components, FIS1, BAK, MIRO, as well as VDAC, which may either be degraded through the proteasome or serve as binding partners for p62. p62 may in turn act as an adaptor molecule through direct interaction with LC3 to recruit autophagosomal membranes to the mitochondria. Parkin can also interact with Ambra1, which in turn activates the PI3K complex around mitochondria to facilitate selective mitophagy.
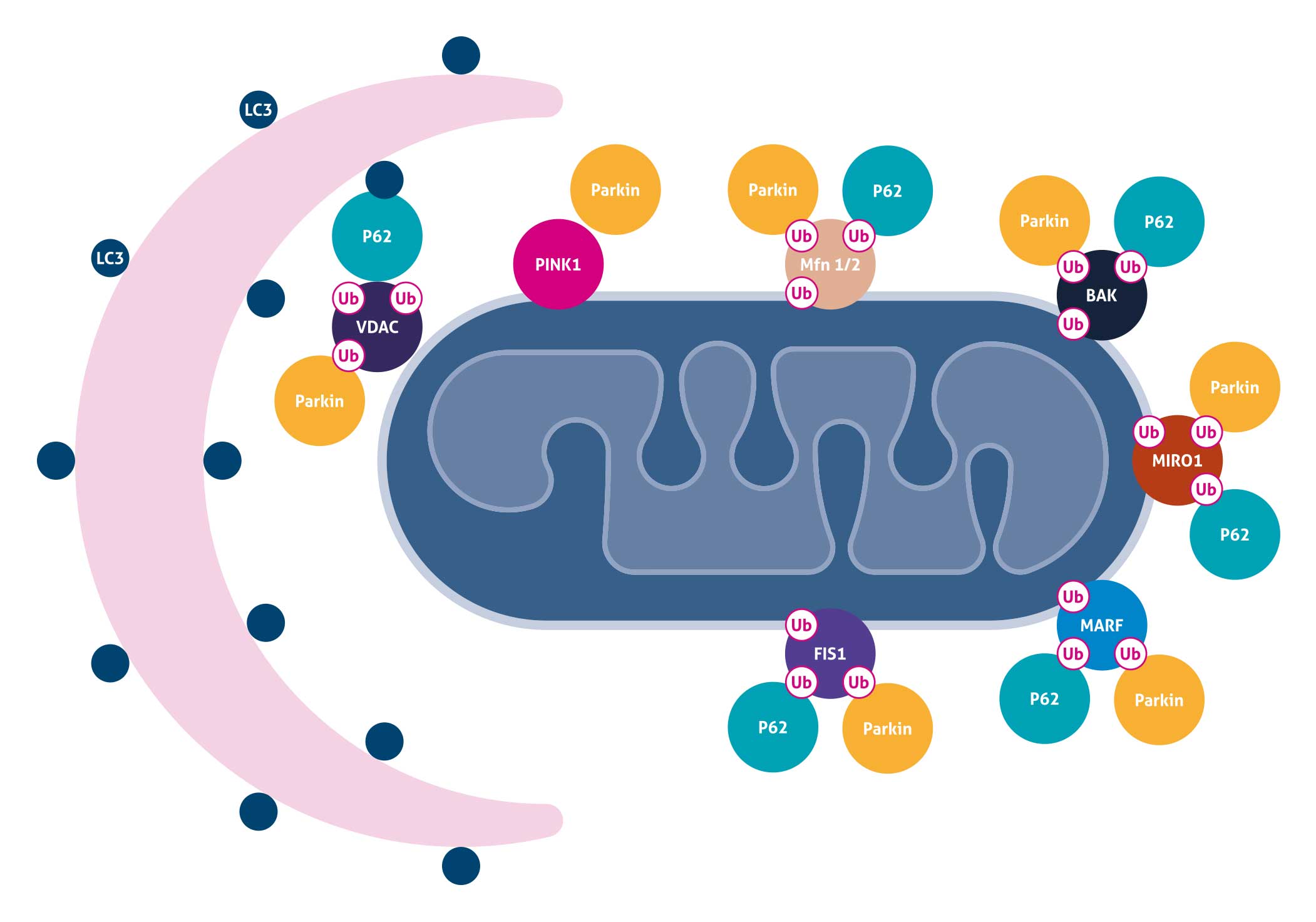
Related antibodies
| Antibody Name | |
| BAK | p62 |
| Beclin 1 | Parkin |
| FIS1 | PINK1 |
| Mfn1 | VDAC1 |
| Mfn2 | VDAC2 |
| NDP52 | VDAC3 |
| OPTN |
Mitochondrial Translation
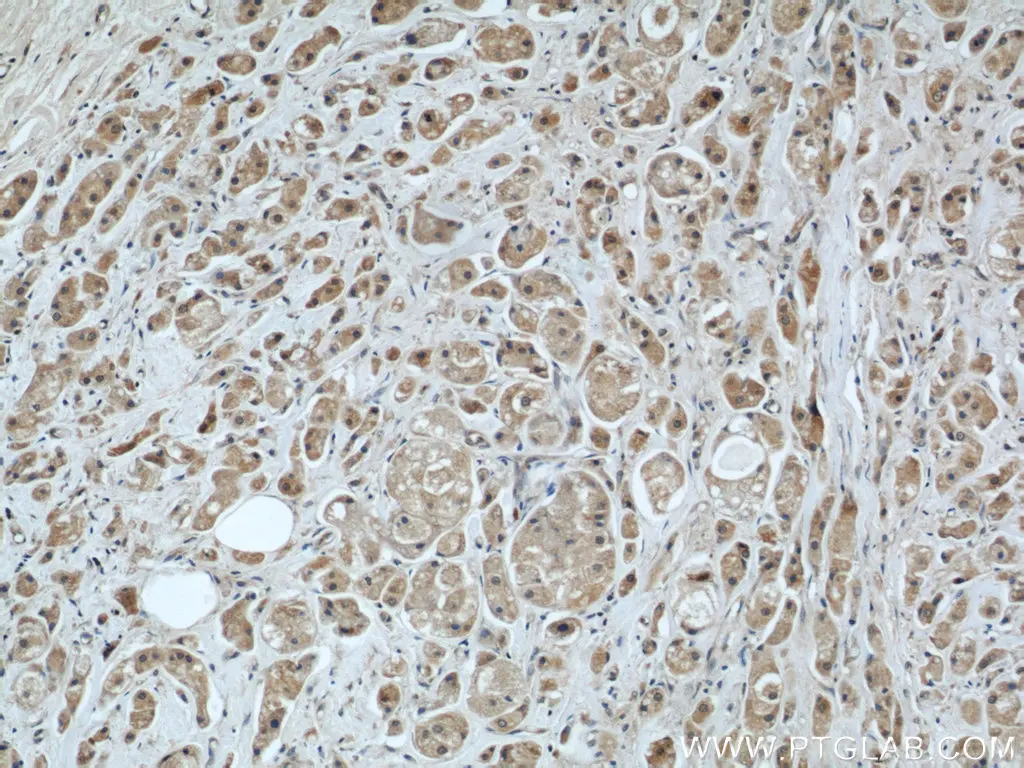
The mitochondria contain their own DNA separated from the nucleus. During the course of evolution, most of the mitochondrial protein-coding genes have been transferred to the nuclear genome. However, a few genes have been retained in the genome of the modern organelle.We now know that the mitochondrial genome (mtDNA), which is housed in the mitochondrial matrix, contains the blueprint for thirteen proteins and all the RNA molecules believed to be necessary and sufficient for intra-mitochondrial protein synthesis. All the other required components for intra-mitochondrial protein synthesis are imported from the cytosol after their synthesis in the cytoplasm. Central to the process of mitochondrial protein synthesis is the mitochondrial ribosome.
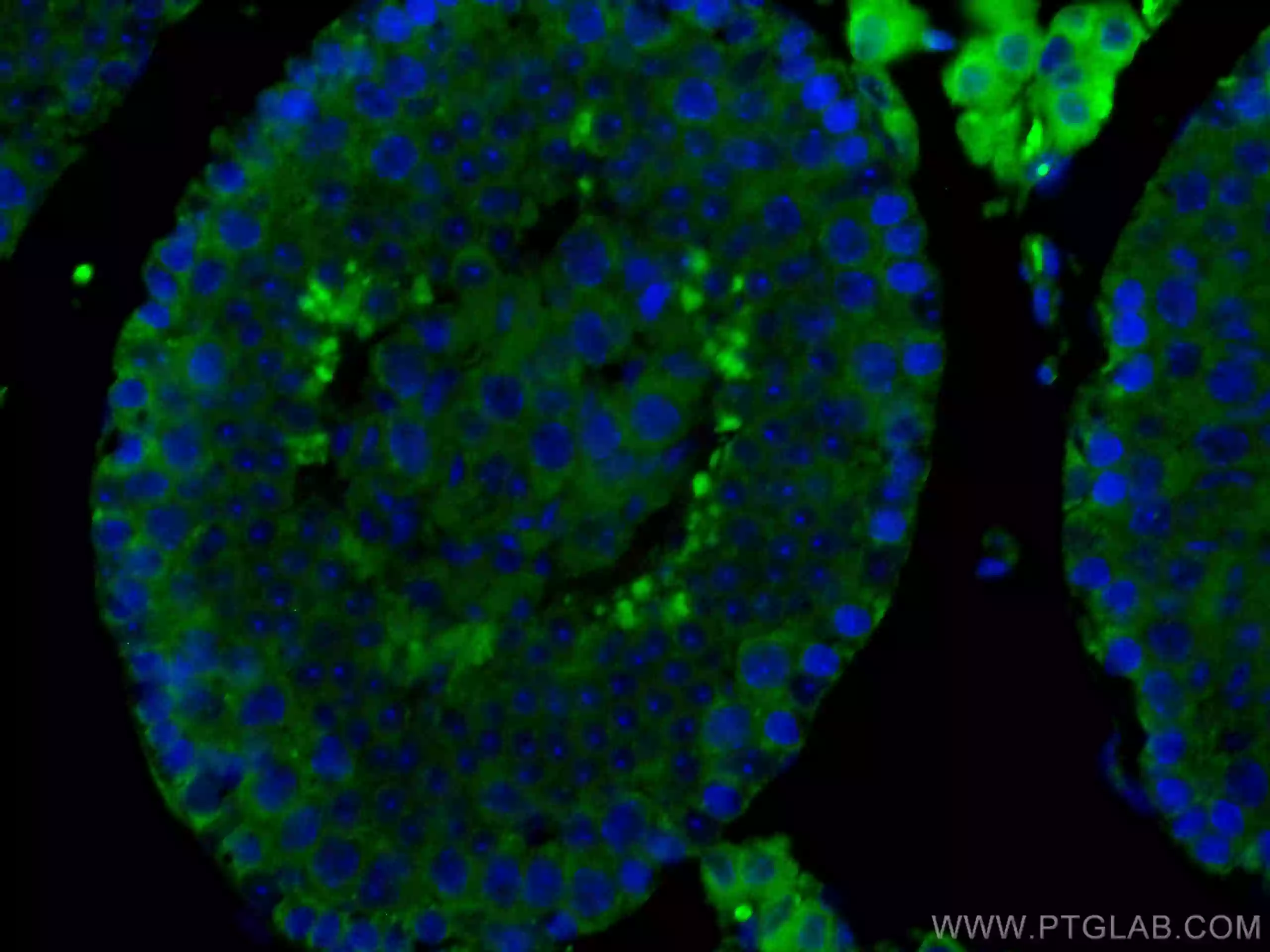
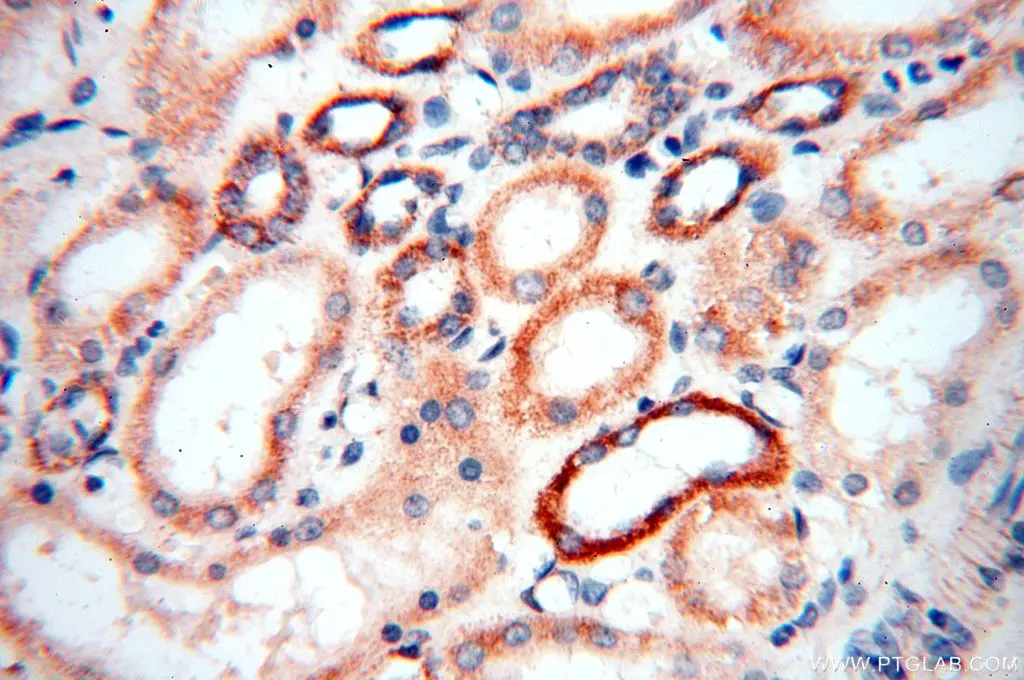
Mitochondrial Protein Import
Though mitochondria possess their own genome and translation machinery, only a small number of mitochondrial proteins, including a few core constituents of the respiratory chain complexes, are encoded by mtDNA and synthesized within the organelle. Thus, the vast majority of mitochondrial proteins are nuclear-encoded and have to be imported into the organelle.Upon synthesis on free ribosomes, mitochondrial precursor proteins reach the surface of the organelle in a process that is guided by cytosolic chaperones. Subsequently, they are imported by specialized protein import machineries and sorted to the designated submitochondrial destination in the outer membrane, inner membrane, intermembrane space, or matrix.
| TOM20 |
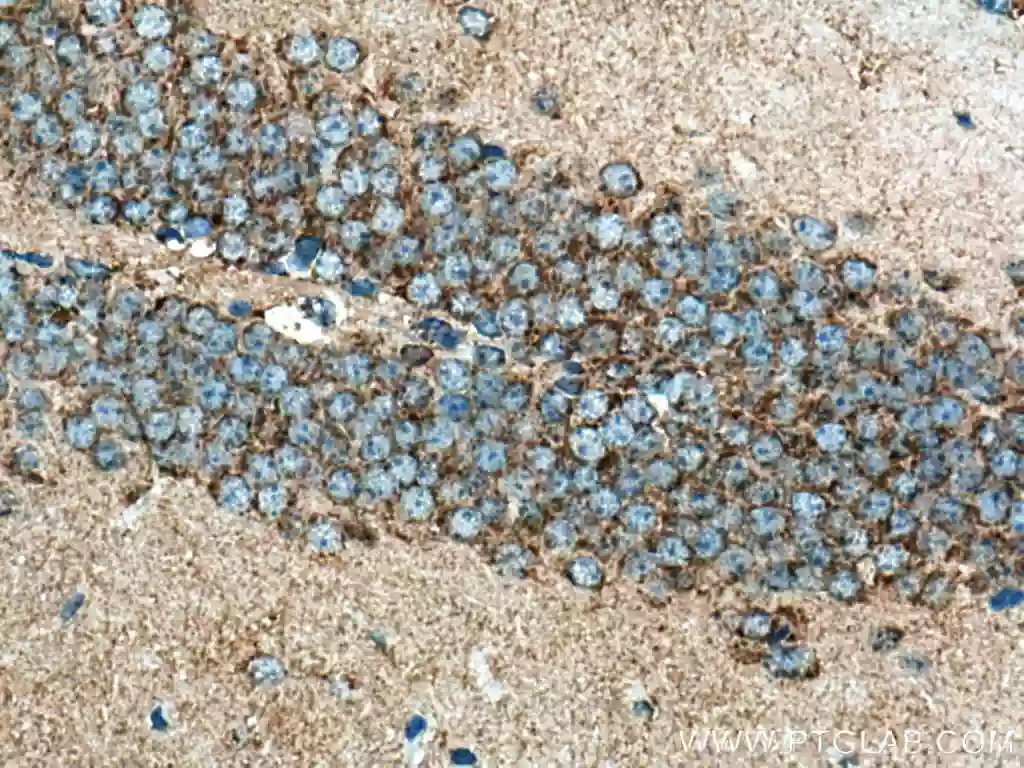 |
| Immunohistochemical analysis of paraffin-embedded mouse brain tissue slide using 11802-1-AP (TOM20 antibody) at dilution of 1:1000 (under 40x lens). Heat mediated antigen retrieval with Tris-EDTA buffer (pH 9.0). |
| TIMM23 |
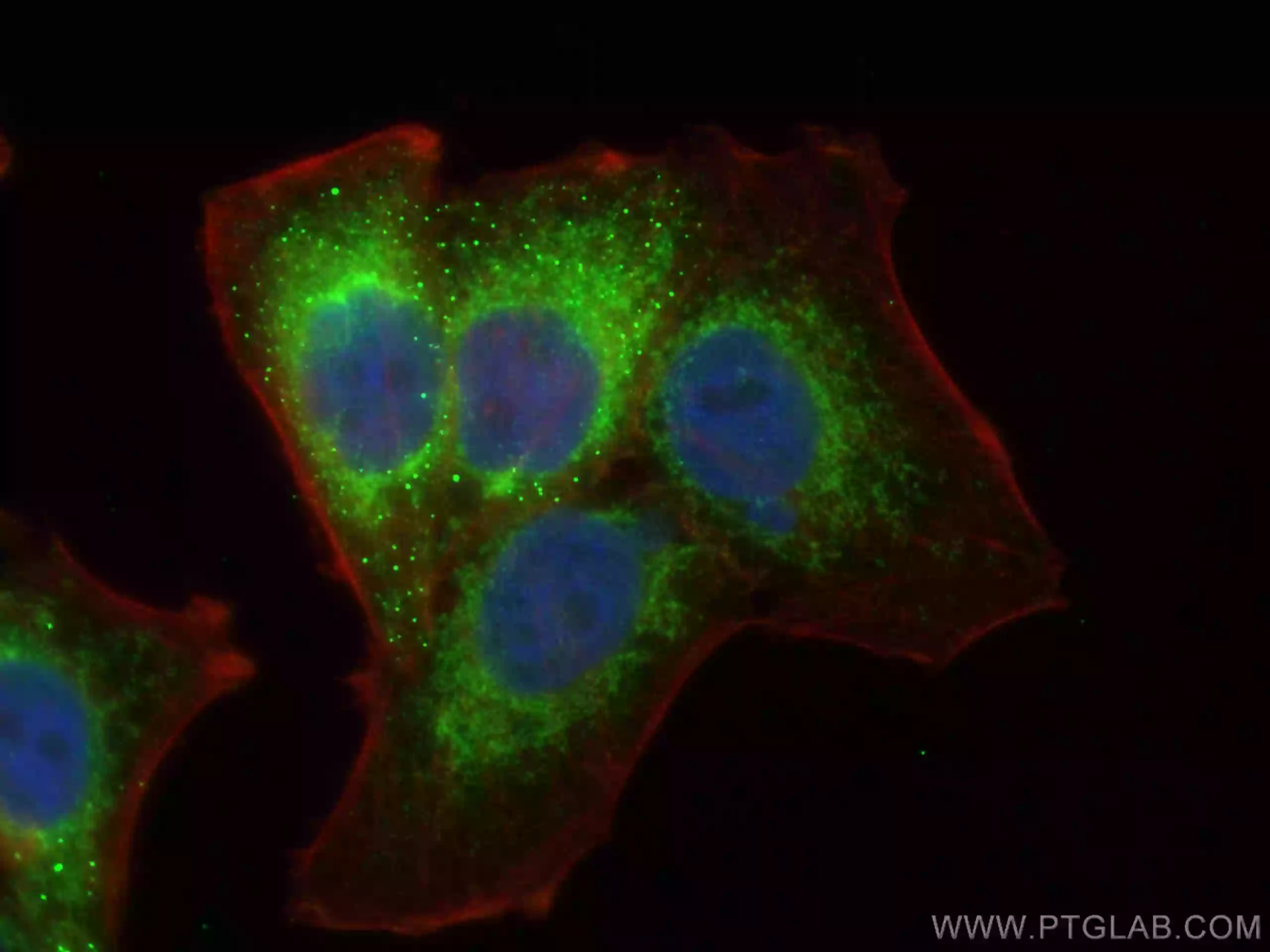 |
|
Immunofluorescent analysis of (4% PFA) fixed HepG2 cells using Tim23 antibody (11123-1-AP) at dilution of 1:200 and CoraLite®488-Conjugated AffiniPure Goat Anti-Rabbit IgG(H+L), CL594-Phallodin (red).
|
| GAPDH Antibody | 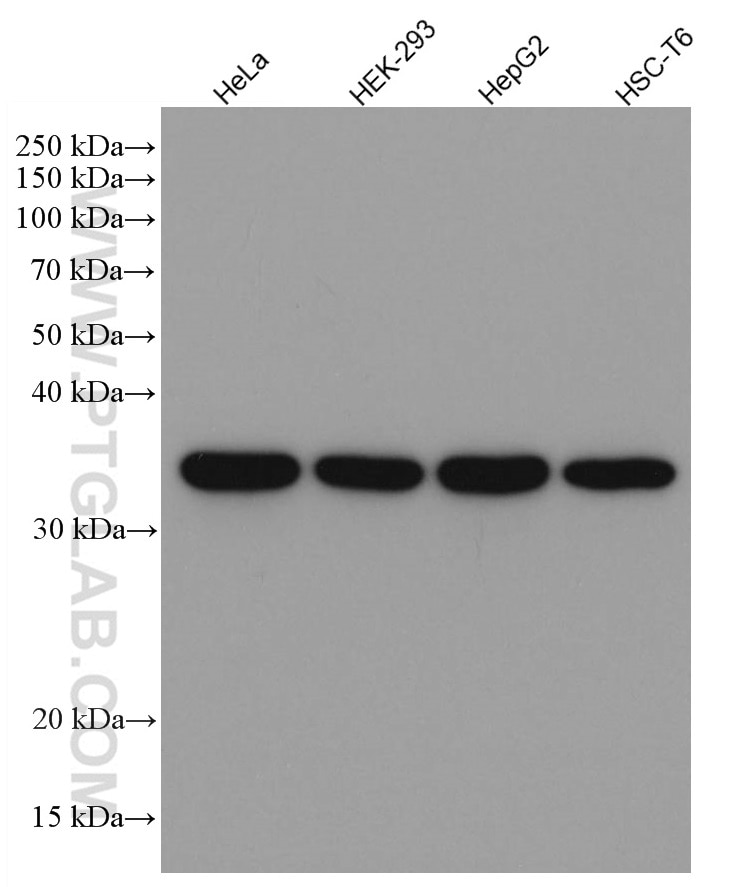 |
| Catalog no.: 60004-1-Ig | |
|
GAPDH is commonly used as a protein loading control in western blot due to its consistently high expression in most cell types. This enzyme participates in several cellular events such as glycolysis, DNA repair, and apoptosis. Proteintech monoclonal GAPDH antibodies are raised against a whole-protein antigen of human origin and have over 4,960 citations. |
| Beta Actin Antibody (KD/KO validated) | 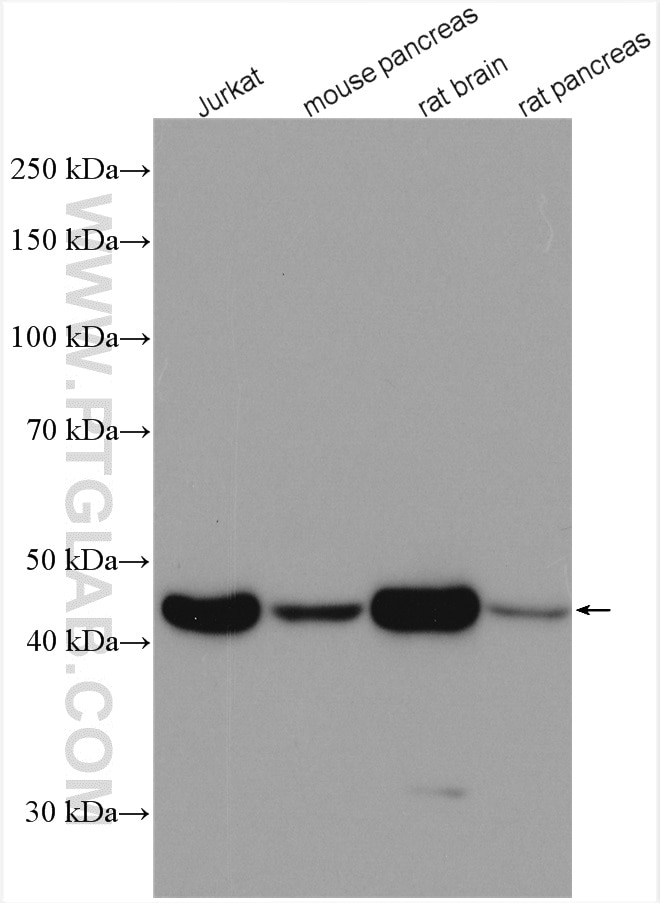 |
| Catalog no.: 66009-1-Ig | |
|
Beta-actin is usually used as a loading control due to its broad and consistent expression across all eukaryotic cell types and the fact that expression levels of this protein are not affected by most experimental treatments. 66009-1-Ig has been cited in over 2,460 publications and has wide species reactivity. |